4LTY
 
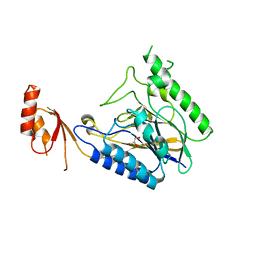 | | Crystal Structure of E.coli SbcD at 1.8 A Resolution | | Descriptor: | Exonuclease subunit SbcD, GLYCEROL | | Authors: | Liu, S, Tian, L.F, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-07-24 | | Release date: | 2014-02-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for DNA recognition and nuclease processing by the Mre11 homologue SbcD in double-strand breaks repair.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7NZN
 
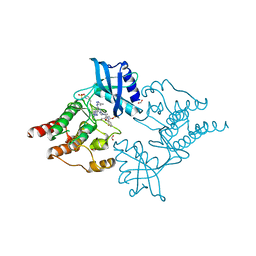 | | Structure of RET kinase domain bound to inhibitor JB-48 | | Descriptor: | 2-[4-[[4-[1-[2-(dimethylamino)ethyl]pyrazol-4-yl]-6-[(3-methyl-1~{H}-pyrazol-5-yl)amino]pyrimidin-2-yl]amino]phenyl]-~{N}-(3-fluorophenyl)ethanamide, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2021-03-24 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of N-Trisubstituted Pyrimidine Derivatives as Type I RET and RET Gatekeeper Mutant Inhibitors with a Novel Kinase Binding Pose.
J.Med.Chem., 65, 2022
|
|
4MRN
 
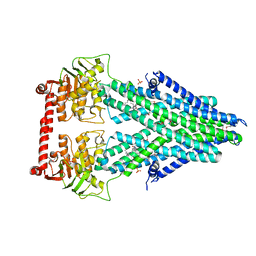 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATE ION | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
4MRS
 
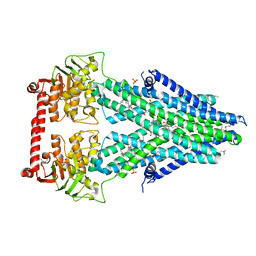 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, OXIDIZED GLUTATHIONE DISULFIDE, ... | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
7OMK
 
 | | The NMR structure of the Zf-GRF domains from the mouse Endonuclease VIII-LIKE 3 (mNEIL3) | | Descriptor: | Endonuclease 8-like 3 | | Authors: | Dinesh, D.C, Huskova, A, Srb, P, Veverka, V, Silhan, J. | | Deposit date: | 2021-05-24 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Model of abasic site DNA cross-link repair; from the architecture of NEIL3 DNA binding domains to the X-structure model.
Nucleic Acids Res., 50, 2022
|
|
1AIJ
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES IN THE CHARGE-NEUTRAL DQAQB STATE | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Stowell, M.H.B, Mcphillips, T.M, Soltis, S.M, Rees, D.C, Abresch, E, Feher, G. | | Deposit date: | 1997-04-18 | | Release date: | 1997-10-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Light-induced structural changes in photosynthetic reaction center: implications for mechanism of electron-proton transfer.
Science, 276, 1997
|
|
1AIG
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES IN THE D+QB-CHARGE SEPARATED STATE | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (II) ION, ... | | Authors: | Stowell, M.H.B, Mcphillips, T.M, Soltis, S.M, Rees, D.C, Abresch, E, Feher, G. | | Deposit date: | 1997-04-17 | | Release date: | 1997-10-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Light-induced structural changes in photosynthetic reaction center: implications for mechanism of electron-proton transfer.
Science, 276, 1997
|
|
4W4I
 
 | |
4W4J
 
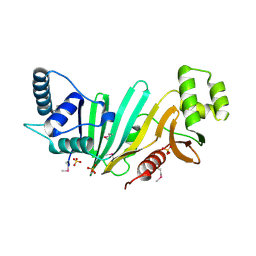 | |
4W4L
 
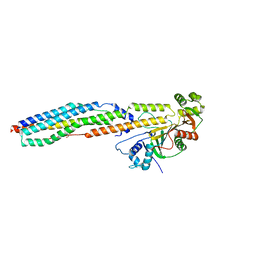 | |
4WES
 
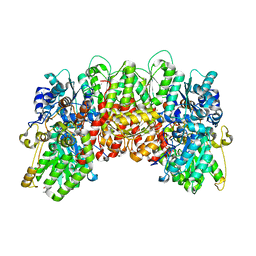 | | Nitrogenase molybdenum-iron protein from Clostridium pasteurianum at 1.08 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (II) ION, ... | | Authors: | Zhang, L.M, Morrison, C.N, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2014-09-10 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Nitrogenase MoFe protein from Clostridium pasteurianum at 1.08 angstrom resolution: comparison with the Azotobacter vinelandii MoFe protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WN9
 
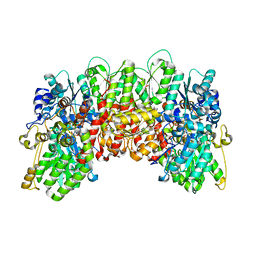 | | Structure of the Nitrogenase MoFe Protein from Clostridium pasteurianum Pressurized with Xenon | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Morrison, C.N, Hoy, J.A, Rees, D.C. | | Deposit date: | 2014-10-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Pathways in the Nitrogenase MoFe Protein by Experimental Identification of Small Molecule Binding Sites.
Biochemistry, 54, 2015
|
|
6ZB9
 
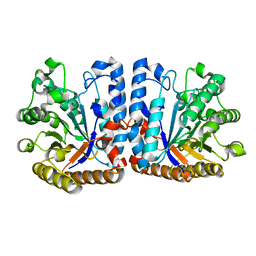 | | Exo-beta-1,3-glucanase from moose rumen microbiome, wild type | | Descriptor: | Exo-beta-1,3-glucanase | | Authors: | Kalyani, D.C, Reichenbach, T, Aspeborg, H, Divne, C. | | Deposit date: | 2020-06-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A homodimeric bacterial exo-beta-1,3-glucanase derived from moose rumen microbiome shows a structural framework similar to yeast exo-beta-1,3-glucanases.
Enzyme.Microb.Technol., 143, 2021
|
|
6ZB8
 
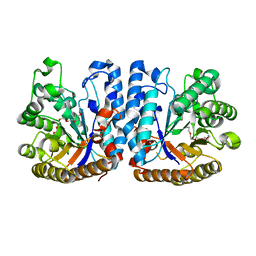 | | Exo-beta-1,3-glucanase from moose rumen microbiome, active site mutant E167Q/E295Q | | Descriptor: | Exo-beta-1,3-glucanase variant E167Q/E295Q, POLYETHYLENE GLYCOL (N=34) | | Authors: | Kalyani, D.C, Reichenbach, T, Aspeborg, H, Divne, C. | | Deposit date: | 2020-06-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A homodimeric bacterial exo-beta-1,3-glucanase derived from moose rumen microbiome shows a structural framework similar to yeast exo-beta-1,3-glucanases.
Enzyme.Microb.Technol., 143, 2021
|
|
4WNA
 
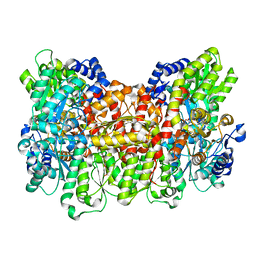 | | Structure of the Nitrogenase MoFe Protein from Azotobacter vinelandii Pressurized with Xenon | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Morrison, C.N, Hoy, J.A, Zhang, L, Einsle, O, Rees, D.C. | | Deposit date: | 2014-10-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Pathways in the Nitrogenase MoFe Protein by Experimental Identification of Small Molecule Binding Sites.
Biochemistry, 54, 2015
|
|
4WOY
 
 | | Crystal structure and functional analysis of MiD49, a receptor for the mitochondrial fission protein Drp1 | | Descriptor: | Mitochondrial dynamics protein MID49 | | Authors: | Loson, O.C, Meng, S, Ngo, H.B, Liu, R, Kaiser, J.T, Chan, D.C. | | Deposit date: | 2014-10-17 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and functional analysis of MiD49, a receptor for the mitochondrial fission protein Drp1.
Protein Sci., 24, 2015
|
|
4WZA
 
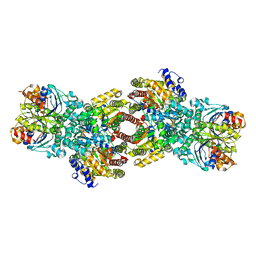 | | Asymmetric Nucleotide Binding in the Nitrogenase Complex | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, ... | | Authors: | Tezcan, F.A, Kaiser, J.T, Howard, J.B, Rees, D.C. | | Deposit date: | 2014-11-19 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8995 Å) | | Cite: | Structural evidence for asymmetrical nucleotide interactions in nitrogenase.
J.Am.Chem.Soc., 137, 2015
|
|
6YXE
 
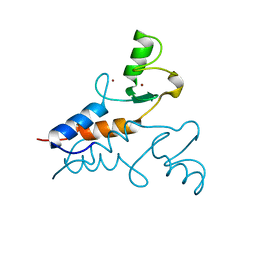 | | Structure of the Trim69 RING domain | | Descriptor: | E3 ubiquitin-protein ligase TRIM69, ZINC ION | | Authors: | Keown, J.R, Goldstone, D.C. | | Deposit date: | 2020-05-01 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The RING domain of TRIM69 promotes higher-order assembly.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4RCR
 
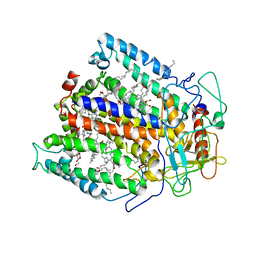 | | STRUCTURE OF THE REACTION CENTER FROM RHODOBACTER SPHAEROIDES R-26 AND 2.4.1: PROTEIN-COFACTOR (BACTERIOCHLOROPHYLL, BACTERIOPHEOPHYTIN, AND CAROTENOID) INTERACTIONS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Komiya, H, Yeates, T.O, Chirino, A.J, Rees, D.C, Allen, J.P, Feher, G. | | Deposit date: | 1991-09-09 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4.1: protein-cofactor (bacteriochlorophyll, bacteriopheophytin, and carotenoid) interactions.
Proc.Natl.Acad.Sci.USA, 85, 1988
|
|
6FPZ
 
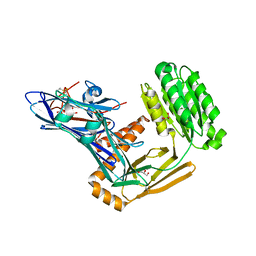 | | Inter-alpha-inhibitor heavy chain 1, D298A | | Descriptor: | ACETATE ION, GLYCEROL, Inter-alpha-trypsin inhibitor heavy chain H1 | | Authors: | Briggs, D.C, Day, A.J. | | Deposit date: | 2018-02-12 | | Release date: | 2019-02-27 | | Last modified: | 2020-03-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inter-alpha-inhibitor heavy chain-1 has an integrin-like 3D structure mediating immune regulatory activities and matrix stabilization during ovulation
J.Biol.Chem., 2020
|
|
1ZUT
 
 | | Crystal Structure Of Mutant K8DP9SR58K Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I, SULFATE ION | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
1GCB
 
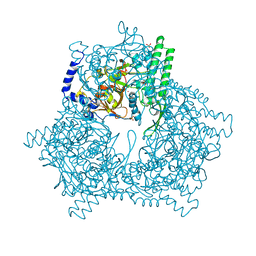 | | GAL6, YEAST BLEOMYCIN HYDROLASE DNA-BINDING PROTEASE (THIOL) | | Descriptor: | GAL6 HG (EMTS) DERIVATIVE, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Joshua-Tor, L, Xu, H.E, Johnston, S.A, Rees, D.C. | | Deposit date: | 1995-07-18 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a conserved protease that binds DNA: the bleomycin hydrolase, Gal6.
Science, 269, 1995
|
|
6P5A
 
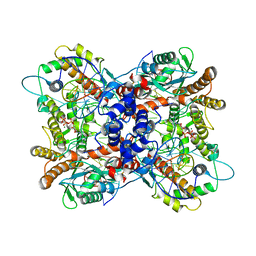 | | Drosophila P element transposase strand transfer complex | | Descriptor: | DNA (44-MER), DNA (5'-D(P*AP*GP*GP*TP*GP*GP*TP*CP*CP*CP*GP*TP*CP*GP*G)-3'), DNA (5'-D(P*CP*GP*AP*AP*CP*TP*AP*TP*A)-3'), ... | | Authors: | Kellogg, E.H, Nogales, E, Ghanim, G, Rio, D.C. | | Deposit date: | 2019-05-30 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a P element transposase-DNA complex reveals unusual DNA structures and GTP-DNA contacts.
Nat.Struct.Mol.Biol., 26, 2019
|
|
1ZYV
 
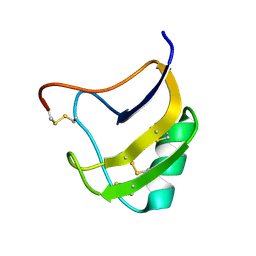 | | Crystal Structure Of Mutant K8DP9SR58KV59G Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-06-12 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
1ZU3
 
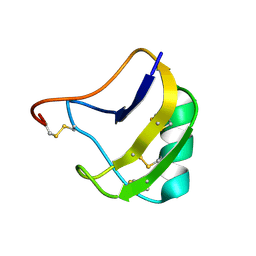 | | Crystal Structure Of Mutant K8A Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-05-30 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
