1XZ0
 
 | | Crystal structure of CD1a in complex with a synthetic mycobactin lipopeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(HYDROXY-HEXADECANOYL-AMINO)-2-{[(4S)-2-(2-HYDROXY-PHENYL)-4,5-DIHYDRO-OXAZOLE-4-CARBONYL]-AMINO}-HEXANOIC ACID 2-[(3S)-1-(TERT-BUTYL-DIPHENYL-SILANYLOXY)-2-OXO-AZEPAN-3-YLCARBAMOYL]-(1S)-1-METHYL-ETHYL ESTER, Beta-2-microglobulin, ... | | Authors: | Zajonc, D.M, Crispin, M.D, Bowden, T.A, Young, D.C, Cheng, T.Y, Hu, J, Costello, C.E, Miller, M.J, Moody, D.B, Wilson, I.A. | | Deposit date: | 2004-11-11 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Mechanism of Lipopeptide Presentation by CD1a.
Immunity, 22, 2005
|
|
1ZJ6
 
 | | Crystal structure of human ARL5 | | Descriptor: | ADP-ribosylation factor-like protein 5, GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Wang, Z.X, Shi, L, Liu, J.F, An, X.M, Chang, W.R, Liang, D.C. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A crystal structure of human ARL5-GDP3'P, a novel member of the small GTP-binding proteins
Biochem.Biophys.Res.Commun., 332, 2005
|
|
1XWE
 
 | | NMR Structure of C345C (NTR) domain of C5 of complement | | Descriptor: | Complement C5 | | Authors: | Bramham, J, Thai, C.-T, Soares, D.C, Uhrin, D, Ogata, R.T, Barlow, P.N. | | Deposit date: | 2004-10-30 | | Release date: | 2004-12-21 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Functional Insights from the Structure of the Multifunctional C345C Domain of C5 of Complement
J.Biol.Chem., 280, 2005
|
|
1ZO9
 
 | | Crystal Structure Of The Wild Type Heme Domain Of P450BM-3 with N-palmitoylmethionine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450:NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Hegda, A, Chen, B, Tomchick, D.R, Bondlela, M, Haines, D.C, Schaffer, N, Machius, M, Graham, S.E, Peterson, J.A. | | Deposit date: | 2005-05-12 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interactions of substrates at the surface of P450s can greatly enhance substrate potency.
Biochemistry, 46, 2007
|
|
1ZHQ
 
 | | Crystal structure of apo MVL | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, mannan-binding lectin | | Authors: | Williams, D.C, Lee, J.Y, Cai, M, Bewley, C.A, Clore, G.M. | | Deposit date: | 2005-04-26 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of the HIV-1 Inhibitory Cyanobacterial Protein MVL Free and Bound to Man3GlcNAc2: STRUCTURAL BASIS FOR SPECIFICITY AND HIGH-AFFINITY BINDING TO THE CORE PENTASACCHARIDE FROM N-LINKED OLIGOMANNOSIDE.
J.Biol.Chem., 280, 2005
|
|
1ZUT
 
 | | Crystal Structure Of Mutant K8DP9SR58K Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I, SULFATE ION | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
1ZXH
 
 | | G311 mutant protein | | Descriptor: | Immunoglobulin G binding protein G | | Authors: | He, Y, Yeh, D.C, Alexander, P, Bryan, P.N, Orban, J. | | Deposit date: | 2005-06-08 | | Release date: | 2005-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of IgG binding domains with artificially evolved high levels of sequence identity but different folds.
Biochemistry, 44, 2005
|
|
1XA9
 
 | | Crystal structure of yellow fluorescent protein zFP538 K66M green mutant | | Descriptor: | BETA-MERCAPTOETHANOL, fluorescent protein FP538 | | Authors: | Remington, S.J, Wachter, R.M, Yarbrough, D.K, Branchaud, B, Anderson, D.C, Kallio, K, Lukyanov, K.A. | | Deposit date: | 2004-08-25 | | Release date: | 2005-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | zFP538, a yellow-fluorescent protein from Zoanthus, contains a novel three-ring chromophore.
Biochemistry, 44, 2005
|
|
1ZYW
 
 | | Crystal Structure Of Mutant K8DP9SR58KP60G Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | ACETATE ION, Alpha-like neurotoxin BmK-I, SULFATE ION | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-06-13 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
1ZYV
 
 | | Crystal Structure Of Mutant K8DP9SR58KV59G Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-06-12 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
1ZU3
 
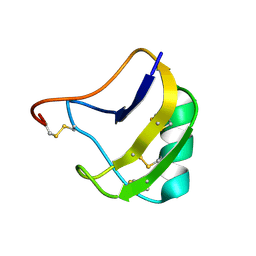 | | Crystal Structure Of Mutant K8A Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-05-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
2AFI
 
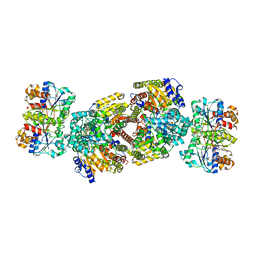 | | Crystal Structure of MgADP bound Av2-Av1 Complex | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Tezcan, F.A, Kaiser, J.T, Mustafi, D, Walton, M.Y, Howard, J.B, Rees, D.C. | | Deposit date: | 2005-07-25 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nitrogenase Complexes: Multiple Docking Sites for a Nucleotide Switch Protein
Science, 309, 2005
|
|
1ZVE
 
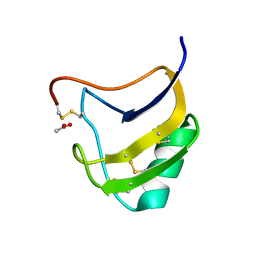 | | Crystal Structure Of Mutant K8G Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | ACETATE ION, Alpha-like neurotoxin BmK-I | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-06-01 | | Release date: | 2006-05-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
1ZVG
 
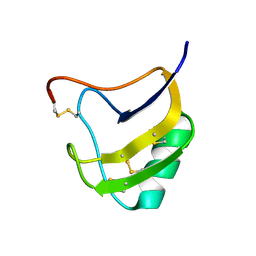 | | Crystal Structure Of Mutant K8DP9S Of Scorpion alpha-Like Neurotoxin Bmk M1 From Buthus Martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Ye, X, Bosmans, F, Li, C, Zhang, Y, Wang, D.C, Tytgat, J. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for the voltage-gated Na+ channel selectivity of the scorpion alpha-like toxin BmK M1
J.Mol.Biol., 353, 2005
|
|
1ZXG
 
 | | Solution structure of A219 | | Descriptor: | Immunoglobulin G binding protein A | | Authors: | He, Y, Yeh, D.C, Alexander, P, Bryan, P.N, Orban, J. | | Deposit date: | 2005-06-08 | | Release date: | 2005-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of IgG binding domains with artificially evolved high levels of sequence identity but different folds.
Biochemistry, 44, 2005
|
|
1SEB
 
 | | COMPLEX OF THE HUMAN MHC CLASS II GLYCOPROTEIN HLA-DR1 AND THE BACTERIAL SUPERANTIGEN SEB | | Descriptor: | ENDOGENOUS PEPTIDE MODEL, POLY-ALA, ENTEROTOXIN TYPE B, ... | | Authors: | Jardetzky, T.S, Brown, J.H, Gorga, J.C, Stern, L.J, Urban, R.G, Chi, Y.I, Stauffacher, C, Strominger, J.L, Wiley, D.C. | | Deposit date: | 1995-11-26 | | Release date: | 1996-06-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of a human class II histocompatibility molecule complexed with superantigen.
Nature, 368, 1994
|
|
1HTM
 
 | | STRUCTURE OF INFLUENZA HAEMAGGLUTININ AT THE PH OF MEMBRANE FUSION | | Descriptor: | HEMAGGLUTININ HA1 CHAIN, HEMAGGLUTININ HA2 CHAIN | | Authors: | Bullough, P.A, Hughson, F.M, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1994-11-02 | | Release date: | 1995-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of influenza haemagglutinin at the pH of membrane fusion.
Nature, 371, 1994
|
|
1SN1
 
 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M1 | | Descriptor: | PROTEIN (NEUROTOXIN BMK M1) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
1M2D
 
 | | Crystal structure at 1.05 Angstroms resolution of the Cys59Ser variant of the thioredoxin-like [2Fe-2S] ferredoxin from Aquifex aeolicus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, [2Fe-2S] ferredoxin | | Authors: | Yeh, A.P, Ambroggio, X.I, Andrade, S.L.A, Einsle, O, Chatelet, C, Meyer, J, Rees, D.C. | | Deposit date: | 2002-06-22 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | High-resolution crystal structures
of the wild type and Cys-55-->Ser and
Cys-59-->Ser variants of the thioredoxin-like
[2Fe-2S] ferredoxin from Aquifex aeolicus
J.Biol.Chem., 277, 2002
|
|
1O2F
 
 | | COMPLEX OF ENZYME IIAGLC AND IIBGLC PHOSPHOCARRIER PROTEIN HPR FROM ESCHERICHIA COLI NMR, RESTRAINED REGULARIZED MEAN STRUCTURE | | Descriptor: | PHOSPHITE ION, PTS system, glucose-specific IIA component, ... | | Authors: | Clore, G.M, Cai, M, Williams, D.C. | | Deposit date: | 2003-03-11 | | Release date: | 2003-05-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Phosphoryl Transfer Complex between the Signal-transducing Protein IIAGlucose and the Cytoplasmic Domain of the Glucose Transporter IICBGlucose of the Escherichia coli Glucose Phosphotransferase System.
J.Biol.Chem., 278, 2003
|
|
1NIP
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF THE NITROGENASE IRON PROTEIN FROM AZOTOBACTER VINELANDII | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Komiya, H, Georgiadis, M.M, Chakrabarti, P, Woo, D, Kornuc, J.J, Rees, D.C. | | Deposit date: | 1992-09-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii.
Science, 257, 1992
|
|
1PKS
 
 | | STRUCTURE OF THE PI3K SH3 DOMAIN AND ANALYSIS OF THE SH3 FAMILY | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Koyama, S, Yu, H, Dalgarno, D.C, Shin, T.B, Zydowsky, L.D, Schreiber, S.L. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the PI3K SH3 domain and analysis of the SH3 family.
Cell(Cambridge,Mass.), 72, 1993
|
|
1PKT
 
 | | STRUCTURE OF THE PI3K SH3 DOMAIN AND ANALYSIS OF THE SH3 FAMILY | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN | | Authors: | Koyama, S, Yu, H, Dalgarno, D.C, Shin, T.B, Zydowsky, L.D, Schreiber, S.L. | | Deposit date: | 1994-03-07 | | Release date: | 1994-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the PI3K SH3 domain and analysis of the SH3 family.
Cell(Cambridge,Mass.), 72, 1993
|
|
1RFA
 
 | | NMR SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF C-RAF-1 | | Descriptor: | RAF1 | | Authors: | Emerson, S.D, Madison, V.S, Palermo, R.E, Waugh, D.S, Scheffler, J.E, Tsao, K.-L, Kiefer, S.E, Liu, S.P, Fry, D.C. | | Deposit date: | 1995-04-26 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of c-Raf-1 and identification of its Ras interaction surface.
Biochemistry, 34, 1995
|
|
1P9Z
 
 | | The Solution Structure of Antifungal Peptide Distinct With a Five-disulfide Motif from Eucommia ulmoides Oliver | | Descriptor: | Eucommia Antifungal peptide 2 | | Authors: | Huang, R.H, Xiang, Y, Tu, G.Z, Zhang, Y, Wang, D.C. | | Deposit date: | 2003-05-13 | | Release date: | 2004-05-25 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Eucommia Antifungal Peptide: A Novel Structural Model Distinct with a Five-Disulfide Motif.
Biochemistry, 43, 2004
|
|
