8CR0
 
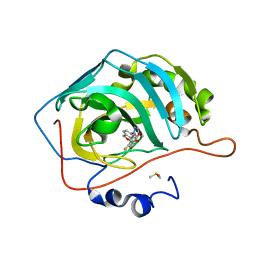 | | Crystal structure of human carbonic anhydrase II in complex with a triazolyl benzoxaborole inhibitor | | Descriptor: | 1-[1,1-bis(oxidanyl)-3~{H}-2,1$l^{4}-benzoxaborol-6-yl]-4-[(4-fluoranylphenoxy)methyl]-1,2,3-triazole, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | 6-Substituted Triazolyl Benzoxaboroles as Selective Carbonic Anhydrase Inhibitors: In Silico Design, Synthesis, and X-ray Crystallography.
J.Med.Chem., 66, 2023
|
|
8CHX
 
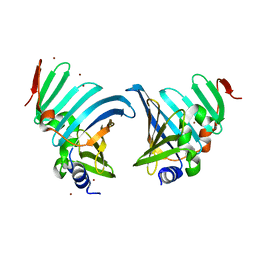 | |
8CR4
 
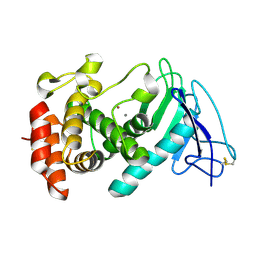 | |
5MMZ
 
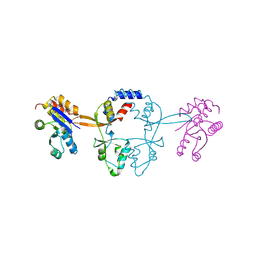 | | Structure of PRL-1 in complex with the Bateman domain of CNNM2 | | Descriptor: | Metal transporter CNNM2, Protein tyrosine phosphatase type IVA 1 | | Authors: | Gimenez-Mascarell, P, Oyenarte, I, Hardy, S, Breiderhoff, T, Stuiver, M, Kostantin, E, Diercks, T, Pey, A.L, Ereno-Orbea, J, Martinez-Chantar, M.L, Khalaf-Nazzal, R, Claverie-Martin, F, Muller, D, Tremblay, M.L, Martinez-Cruz, L.A. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of the Oncogenic Interaction of Phosphatase PRL-1 with the Magnesium Transporter CNNM2.
J. Biol. Chem., 292, 2017
|
|
8CGM
 
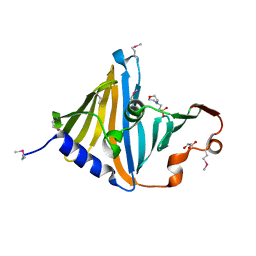 | | Structure of the lipoprotein transporter LolA from Porphyromonas gingivalis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Persson, K, Jaiman, D, Nagampalli, R. | | Deposit date: | 2023-02-06 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A comparative analysis of lipoprotein transport proteins: LolA and LolB from Vibrio cholerae and LolA from Porphyromonas gingivalis.
Sci Rep, 13, 2023
|
|
5MN4
 
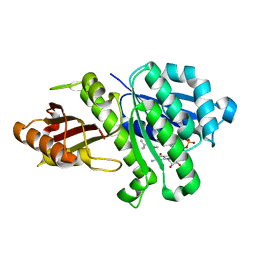 | | S. aureus FtsZ 12-316 F138A GDP Open form (1FOf) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Wagstaff, J.M, Tsim, M, Kureisaite-Ciziene, D, Lowe, J. | | Deposit date: | 2016-12-12 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Polymerization-Associated Structural Switch in FtsZ That Enables Treadmilling of Model Filaments.
MBio, 8, 2017
|
|
8CM1
 
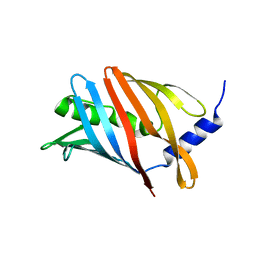 | | Lol B - Localization of lipoprotein B from Vibrio cholera | | Descriptor: | Outer-membrane lipoprotein LolB | | Authors: | Jaiman, D, Persson, K. | | Deposit date: | 2023-02-17 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A comparative analysis of lipoprotein transport proteins: LolA and LolB from Vibrio cholerae and LolA from Porphyromonas gingivalis.
Sci Rep, 13, 2023
|
|
5MMM
 
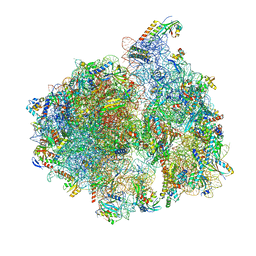 | | Structure of the 70S chloroplast ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein 2, ... | | Authors: | Bieri, P, Leibundgut, M, Saurer, M, Boehringer, D, Ban, N. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The complete structure of the chloroplast 70S ribosome in complex with translation factor pY.
EMBO J., 36, 2017
|
|
8CPH
 
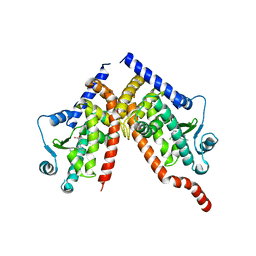 | | Crystal structure of PPAR gamma (PPARG) in complex with WY-14643 (inactive form) | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
8CPI
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with WY-14643 | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
8CPJ
 
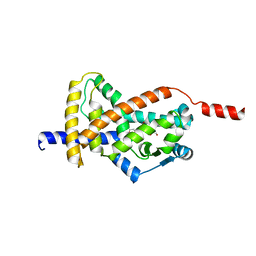 | | Crystal structure of PPAR gamma (PPARG) in an inactive form | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
5MRG
 
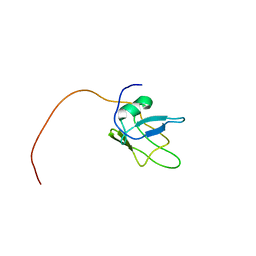 | | Solution structure of TDP-43 (residues 1-102) | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Mompean, M, Romano, V, Pantoja-Uceda, D, Stuani, C, Baralle, F.E, Laurents, D.V. | | Deposit date: | 2016-12-22 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Point mutations in the N-terminal domain of transactive response DNA-binding protein 43 kDa (TDP-43) compromise its stability, dimerization, and functions.
J. Biol. Chem., 292, 2017
|
|
5MA3
 
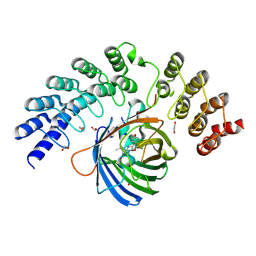 | | GFP-binding DARPin fusion gc_R11 | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein, R11 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
5MSY
 
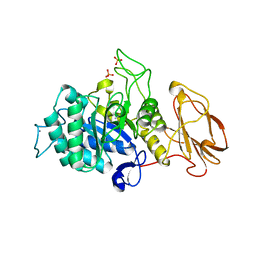 | | Glycoside hydrolase BT_1012 | | Descriptor: | AMMONIA, Glycoside hydrolase, PHOSPHATE ION | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
7AE7
 
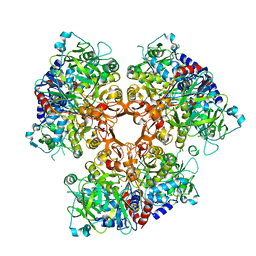 | |
7AE4
 
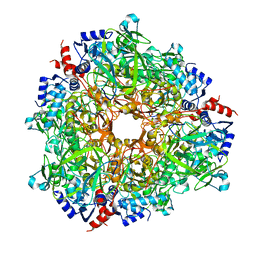 | |
8C29
 
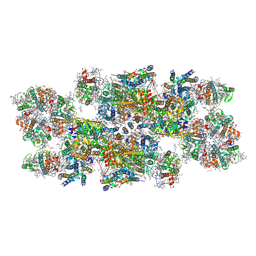 | | Cryo-EM structure of photosystem II C2S2 supercomplex from Norway spruce (Picea abies) at 2.8 Angstrom resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Kopecny, D, Semchonok, D.A, Kouril, R. | | Deposit date: | 2022-12-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.785 Å) | | Cite: | Cryo-EM structure of a plant photosystem II supercomplex with light-harvesting protein Lhcb8 and alpha-tocopherol.
Nat.Plants, 9, 2023
|
|
5MA5
 
 | | GFP-binding DARPin fusion gc_K11 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Green fluorescent protein, ... | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
7AE5
 
 | |
5MCW
 
 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-LWC2) | | Descriptor: | Cellular tumor antigen p53, DNA, FORMYL GROUP, ... | | Authors: | Golovenko, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2016-11-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
8CG3
 
 | |
3VT0
 
 | | Crystal structure of Ct1,3Gal43A in complex with lactose | | Descriptor: | GLYCEROL, Ricin B lectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Jiang, D, Fan, J, Wang, X, Zhao, Y, Huang, B, Zhang, X.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Crystal structure of 1,3Gal43A, an exo-beta-1,3-galactanase from Clostridium thermocellum
J.Struct.Biol., 180, 2012
|
|
5MGV
 
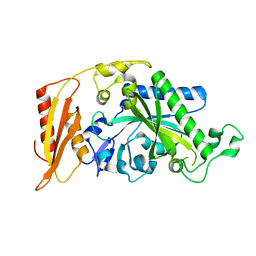 | | Kinetic and Structural Changes in HsmtPheRS, Induced by Pathogenic Mutations in Human FARS2 | | Descriptor: | Phenylalanine--tRNA ligase, mitochondrial | | Authors: | Kartvelishvili, E, Tworowski, D, Vernon, H, Chrzanowska-Lightowlers, Z, Moor, N, Wang, J, Wong, L.-J, Safro, M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Kinetic and structural changes in HsmtPheRS, induced by pathogenic mutations in human FARS2.
Protein Sci., 26, 2017
|
|
7A0S
 
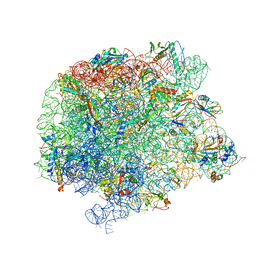 | | 50S Deinococcus radiodurans ribosome bounded with mycinamicin I | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Breiner, E, Eyal, Z, Matzov, D, Halfon, Y, Cimicata, G, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2020-08-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Ribosome-binding and anti-microbial studies of the mycinamicins, 16-membered macrolide antibiotics from Micromonospora griseorubida.
Nucleic Acids Res., 49, 2021
|
|
5MQG
 
 | |
