1YF0
 
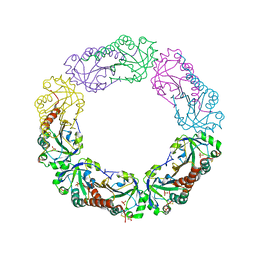 | | Structural and biochemical analysis of the link between enzymatic activity and oligomerization in AhpC, a bacterial peroxiredoxin. | | Descriptor: | Alkyl hydroperoxide reductase subunit C, SULFATE ION | | Authors: | Parsonage, D, Youngblood, D.S, Sarma, G.N, Wood, Z.A, Karplus, P.A, Poole, L.B. | | Deposit date: | 2004-12-29 | | Release date: | 2005-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Analysis of the Link between Enzymatic Activity and Oligomeric State in AhpC, a Bacterial Peroxiredoxin.
Biochemistry, 44, 2005
|
|
6W77
 
 | | 30S-Inactivated-high-Mg2+ Class A | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Jahagirdar, D, Jha, V, Basu, B, Gomez-Blanco, J, Vargas, J, Ortega, J. | | Deposit date: | 2020-03-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Alternative conformations and motions adopted by 30S ribosomal subunits visualized by cryo-electron microscopy.
Rna, 26, 2020
|
|
1YJE
 
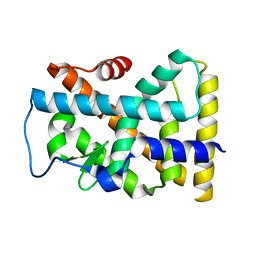 | | Crystal structure of the rNGFI-B ligand-binding domain | | Descriptor: | Orphan nuclear receptor NR4A1 | | Authors: | Flaig, R, Greschik, H, Peluso-Iltis, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the cell-specific activities of the NGFI-B and the Nurr1 ligand-binding domain.
J.Biol.Chem., 280, 2005
|
|
2YQH
 
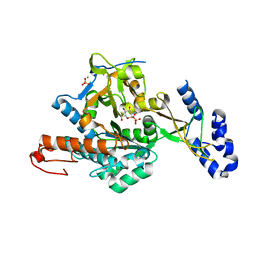 | | Crystal structure of uridine-diphospho-N-acetylglucosamine pyrophosphorylase from Candida albicans, in the substrate-binding form | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Miki, K, Maruyama, D, Nishitani, Y, Nonaka, T, Kita, A. | | Deposit date: | 2007-03-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Uridine-diphospho-N-acetylglucosamine Pyrophosphorylase from Candida albicans and Catalytic Reaction Mechanism
J.Biol.Chem., 282, 2007
|
|
2Y7Z
 
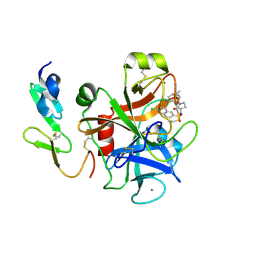 | | Structure and property based design of factor Xa inhibitors: pyrrolidin-2-ones with aminoindane and phenylpyrrolidine P4 motifs | | Descriptor: | 6-CHLORO-N-[(3S)-1-[(1S)-1-DIMETHYLAMINO-2,3-DIHYDRO-1H-INDEN-5-YL]-2-OXO-PYRROLIDIN-3-YL]NAPHTHALENE-2-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Young, R.J, Adams, C, Blows, M, Brown, D, Burns-Kurtis, C.L, Chaudry, L, Chan, C, Convery, M.A, Davies, D.E, Exall, A.M, Foster, G, Harling, J.D, Hortense, E, Irving, W.R, Irvine, S, Jackson, S, Kleanthous, S, Pateman, A.J, Patikis, A.N, Roethka, T.J, Senger, S, Stelman, G.J, Toomey, J.R, West, R.I, Whittaker, C, Zhou, P, Watson, N.S. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-16 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure and Property Based Design of Factor Xa Inhibitors: Pyrrolidin-2-Ones with Aminoindane and Phenylpyrrolidine P4 Motifs.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1YJO
 
 | | Structure of NNQQNY from yeast prion Sup35 with zinc acetate | | Descriptor: | ACETIC ACID, Eukaryotic peptide chain release factor GTP-binding subunit, ZINC ION | | Authors: | Nelson, R, Sawaya, M.R, Balbirnie, M, Madsen, A.O, Riekel, C, Grothe, R, Eisenberg, D. | | Deposit date: | 2005-01-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the cross-beta spine of amyloid-like fibrils.
Nature, 435, 2005
|
|
1YJP
 
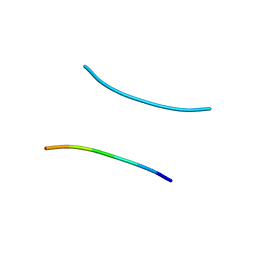 | | Structure of GNNQQNY from yeast prion Sup35 | | Descriptor: | Eukaryotic peptide chain release factor GTP-binding subunit | | Authors: | Nelson, R, Sawaya, M.R, Balbirnie, M, Madsen, A.O, Riekel, C, Grothe, R, Eisenberg, D. | | Deposit date: | 2005-01-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the cross-beta spine of amyloid-like fibrils.
Nature, 435, 2005
|
|
6VOL
 
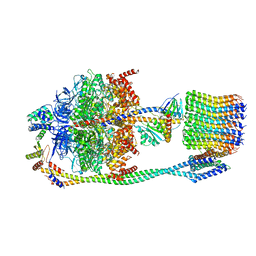 | | Chloroplast ATP synthase (R2, CF1FO) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
1YC0
 
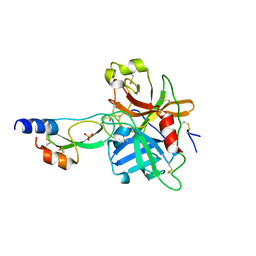 | | short form HGFA with first Kunitz domain from HAI-1 | | Descriptor: | Hepatocyte growth factor activator, Kunitz-type protease inhibitor 1, PHOSPHATE ION | | Authors: | Shia, S, Stamos, J, Kirchhofer, D, Fan, B, Wu, J, Corpuz, R.T, Santell, L, Lazarus, R.A, Eigenbrot, C. | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational lability in serine protease active sites: structures of hepatocyte growth factor activator (HGFA) alone and with the inhibitory domain from HGFA inhibitor-1B.
J.Mol.Biol., 346, 2005
|
|
6VRM
 
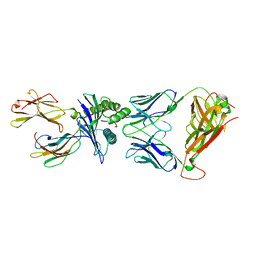 | | T cell receptor-p53-HLA-A2 complex | | Descriptor: | Beta-2-microglobulin, Cellular tumor antigen p53 peptide, MHC class I antigen, ... | | Authors: | Wu, D, Gallagher, D.T, Gowthaman, R, Pierce, B.G, Mariuzza, R.A. | | Deposit date: | 2020-02-08 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Nat Commun, 11, 2020
|
|
2V7U
 
 | |
1YCL
 
 | | Crystal Structure of B. subtilis LuxS in Complex with a Catalytic 2-Ketone Intermediate | | Descriptor: | (S)-2-AMINO-4-[(2S,3R)-2,3,5-TRIHYDROXY-4-OXO-PENTYL]MERCAPTO-BUTYRIC ACID, COBALT (II) ION, S-ribosylhomocysteinase, ... | | Authors: | Rajan, R, Zhu, J, Hu, X, Pei, D, Bell, C.E. | | Deposit date: | 2004-12-22 | | Release date: | 2005-03-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of S-Ribosylhomocysteinase (LuxS) in Complex with a Catalytic 2-Ketone Intermediate.
Biochemistry, 44, 2005
|
|
2UVM
 
 | | Structure of PKBalpha PH domain in complex with a novel inositol headgroup surrogate, benzene 1,2,3,4-tetrakisphosphate | | Descriptor: | BENZENE-1,2,3,4-TETRAYL TETRAKIS[DIHYDROGEN (PHOSPHATE)], RAC-ALPHA SERINE/THREONINE-PROTEIN KINASE, SODIUM ION | | Authors: | Komander, D, Mills, S.J, Trusselle, M.N, Safrany, S.T, van Aalten, D.M.F, Potter, B.V.L. | | Deposit date: | 2007-03-12 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel Inositol Phospholipid Headgroup Surrogate Crystallised in the Pleckstrin Homology Domain of Protein Kinase Balpha.
Acs Chem.Biol., 2, 2007
|
|
6VOH
 
 | | Chloroplast ATP synthase (O1, CF1FO) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
6VPY
 
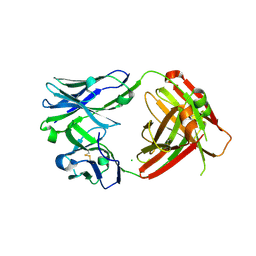 | | I33M (I3.2 mutant from CH103 Lineage) | | Descriptor: | CHLORIDE ION, GLYCEROL, I33M heavy chain, ... | | Authors: | Fera, D, Zhou, J. | | Deposit date: | 2020-02-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The Effects of Framework Mutations at the Variable Domain Interface on Antibody Affinity Maturation in an HIV-1 Broadly Neutralizing Antibody Lineage.
Front Immunol, 11, 2020
|
|
6W1O
 
 | | RT XFEL structure of the dark-stable state of Photosystem II (0F, S1-rich) at 2.08 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VU8
 
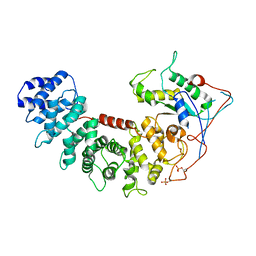 | | Structure of G-alpha-i bound to its chaperone Ric-8A | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1, Resistance to inhibitors of cholinesterase 8 homolog A (C. elegans) | | Authors: | Seven, A.B, Hilger, D. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-18 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structures of G alpha Proteins in Complex with Their Chaperone Reveal Quality Control Mechanisms.
Cell Rep, 30, 2020
|
|
2V40
 
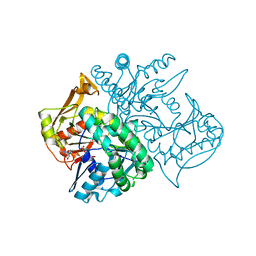 | | Human Adenylosuccinate synthetase isozyme 2 in complex with GDP | | Descriptor: | ADENYLOSUCCINATE SYNTHETASE ISOZYME 2, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Welin, M, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-27 | | Release date: | 2007-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human Adenylosuccinate Synthetase Isozyme 2 in Complex with Gdp
To be Published
|
|
6W2Q
 
 | | Junction 34, DHR53-DHR4 | | Descriptor: | CALCIUM ION, Junction 34 | | Authors: | Bick, M.J, Brunette, T.J, Baker, D. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modular repeat protein sculpting using rigid helical junctions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2VED
 
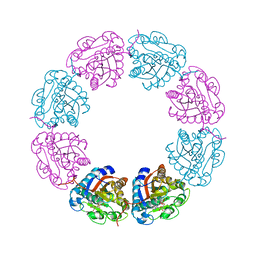 | | crystal structure of the chimerical mutant CapABK55M protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MEMBRANE PROTEIN CAPA1, ... | | Authors: | Olivares-Illana, V, Meyer, P, Gueguen-Chaignon, V, Soulat, D, Deustcher, J, Cozzone, A.J, Morera, S, Grangeasse, C, Nessler, S. | | Deposit date: | 2007-10-19 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Regulation Mechanism of the Tyrosine Kinase Capb from Staphylococcus Aureus.
Plos Biol., 6, 2008
|
|
1YJ1
 
 | | X-ray Crystal Structure of a Chemically Synthesized [D-Gln35]Ubiquitin | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ubiquitin | | Authors: | Bang, D, Makhatadze, G.I, Tereshko, V, Kossiakoff, A.A, Kent, S.B. | | Deposit date: | 2005-01-13 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray Crystal Structure of a Chemically Synthesized [D-Gln35]Ubiquitin
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
2VLQ
 
 | | F86A mutant of E9 DNase domain in complex with Im9 | | Descriptor: | COLICIN E9, COLICIN-E9 IMMUNITY PROTEIN, MALONIC ACID | | Authors: | Keeble, A.H, Joachimiak, L.A, Mate, M.J, Meenan, N, Kirkpatrick, N, Baker, D, Kleanthous, C. | | Deposit date: | 2008-01-15 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Experimental and Computational Analyses of the Energetic Basis for Dual Recognition of Immunity Proteins by Colicin Endonucleases.
J.Mol.Biol., 379, 2008
|
|
6W3D
 
 | | Rd1NTF2_05 with long sheet | | Descriptor: | Rd1NTF2_05 | | Authors: | Bick, M.J, Basanta, B, Sankaran, B, Baker, D. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2UYC
 
 | |
6W3G
 
 | | Rd1NTF2_04 with long sheet | | Descriptor: | Rd1NTF2_04 | | Authors: | Bick, M.J, Basanta, B, Sankaran, B, Baker, D. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | An enumerative algorithm for de novo design of proteins with diverse pocket structures.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
