7LU2
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 6 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7LU3
 
 | | X-ray radiation damage series on Proteinase K at 100K, crystal structure, dataset 7 | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-02-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Evaluating the impact of X-ray damage on conformational heterogeneity in room-temperature (277 K) and cryo-cooled protein crystals.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7M1W
 
 | |
7MBG
 
 | | SARS-CoV-2 Main protease in orthorhombic space group | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Douangamath, A, von Delft, F, Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
7MHN
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 277 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1908 Å) | | Cite: | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
7MHK
 
 | | Crystal Structure of Apo/Unliganded SARS-CoV-2 Main Protease (Mpro) at 310 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9601 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHJ
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 298 K and High Humidity | | Descriptor: | 3C-like proteinase, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.0005 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MNG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor VBY-825 (Partial Occupancy) | | Descriptor: | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name), 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7MHO
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 298 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
7MHI
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 298 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHQ
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 310 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9601 Å) | | Cite: | The temperature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro )
Iucrj, 9, 2022
|
|
7MHH
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 277 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1908 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
8EFB
 
 | | Oliceridine-bound mu-opioid receptor-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EDT
 
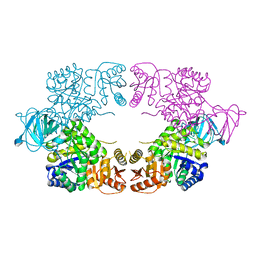 | |
8EFO
 
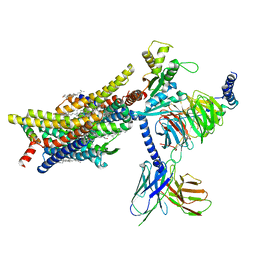 | | PZM21-bound mu-opioid receptor-Gi complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
6QEI
 
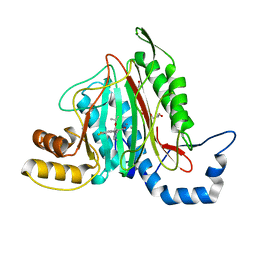 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX WITH AN INHIBITOR 5,6-Difluoro-3-(2-isopropoxy-4-piperazin-1-yl-phenyl)-1H-indole-2-carboxylic acid amide | | Descriptor: | 1,2-ETHANEDIOL, 5,6-bis(fluoranyl)-3-(4-piperazin-1-yl-2-propan-2-yloxy-phenyl)-1~{H}-indole-2-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Musil, D, Heinrich, T, Lehmann, M. | | Deposit date: | 2019-01-07 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Structure-Based Optimization of Next-Generation Reversible Methionine Aminopeptidase-2 (MetAP-2) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6QBO
 
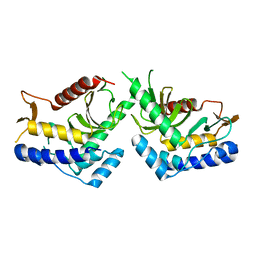 | | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S203A | | Descriptor: | Cell wall assembly regulator SMI1 | | Authors: | Ramos, N, Batista, M, Francois, J.M, Mourey, L, Maveyraud, L, Zerbib, D. | | Deposit date: | 2018-12-21 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | structure of the core domaine of Knr4, an intrinsically disordered protein from Saccharomyces cerevisiae - mutant S203A
To Be Published
|
|
4O32
 
 | | Structure of a malarial protein | | Descriptor: | CHLORIDE ION, Thioredoxin | | Authors: | Egea, P.F, Koehl, A, Peng, M, Cascio, D. | | Deposit date: | 2013-12-17 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Crystal structure and solution characterization of the thioredoxin-2 from Plasmodium falciparum, a constituent of an essential parasitic protein export complex.
Biochem.Biophys.Res.Commun., 456, 2015
|
|
4NYU
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with acetate ion (ACT) in C 2 2 21 | | Descriptor: | ACETATE ION, CALCIUM ION, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8EFQ
 
 | | DAMGO-bound mu-opioid receptor-Gi complex | | Descriptor: | DAMGO, ETHANOLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhuang, Y, Wang, Y, Guo, S, Zhou, X.E, Rao, Q, He, X, He, B, Liu, J, Zhou, Q, Wang, X, Liu, W, Jiang, X, Yang, D, Chen, X, Jiang, Y, Jiang, H, Shen, J, Melcher, K, Wang, M, Xie, X, Xu, H.E. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition of morphine and fentanyl by the human mu-opioid receptor.
Cell, 185, 2022
|
|
8EEX
 
 | | Cas7-11 in complex with Csx29 | | Descriptor: | Cas7-11, Csx29, ZINC ION, ... | | Authors: | Demircioglu, F.E, Wilkinson, M.E, Strecker, J, Li, D, Faure, G, Macrae, R.K, Zhang, F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | RNA-activated protein cleavage with a CRISPR-associated endopeptidase.
Science, 378, 2022
|
|
8E3X
 
 | | Cryo-EM structure of the PAC1R-PACAP27-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Piper, S.J, Danev, R, Sexton, P, Wootten, D. | | Deposit date: | 2022-08-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Understanding VPAC receptor family peptide binding and selectivity.
Nat Commun, 13, 2022
|
|
8EDS
 
 | |
6QEH
 
 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX WITH AN INHIBITOR 5-Chloro-quinolin-8-ol | | Descriptor: | 5-chloranylquinolin-8-ol, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Musil, D, Heinrich, T, Lehmann, M. | | Deposit date: | 2019-01-07 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery and Structure-Based Optimization of Next-Generation Reversible Methionine Aminopeptidase-2 (MetAP-2) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6QJ5
 
 | | X-ray structure of PPARgamma LBD with the ligand NV1380 | | Descriptor: | (2~{S})-3-methyl-2-[(4-octoxyphenyl)carbonylamino]butanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D. | | Deposit date: | 2019-01-22 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Novel N-Substituted Valine Derivative with Unique Peroxisome Proliferator-Activated Receptor gamma Binding Properties and Biological Activities.
J.Med.Chem., 63, 2020
|
|
