7Q1M
 
 | | Crystal structure of human butyrylcholinesterase in complex with N-[(2S)-3-[(cyclohexylmethyl)amino]-2-hydroxypropyl]-2,2-diphenylacetamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Brazzolotto, X, Panek, D, Pasieka, A, Malawska, B, Nachon, F. | | Deposit date: | 2021-10-20 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Discovery of new, highly potent and selective inhibitors of BuChE - design, synthesis, in vitro and in vivo evaluation and crystallography studies.
Eur.J.Med.Chem., 249, 2023
|
|
5LKX
 
 | | Crystal structure of the p300 acetyltransferase catalytic core with propionyl-coenzyme A. | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Histone acetyltransferase p300,Histone acetyltransferase p300, ... | | Authors: | Kaczmarska, Z, Ortega, E, Marquez, J.A, Panne, D. | | Deposit date: | 2016-07-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of p300 in complex with acyl-CoA variants.
Nat. Chem. Biol., 13, 2017
|
|
1Z19
 
 | | Crystal structure of a lambda integrase(75-356) dimer bound to a COC' core site | | Descriptor: | 33-MER, 5'-D(*CP*TP*CP*GP*TP*TP*CP*AP*GP*CP*TP*TP*TP*TP*TP*T)-3', 5'-D(P*TP*TP*TP*AP*TP*AP*CP*TP*AP*AP*GP*TP*TP*GP*GP*CP*AP*TP*TP*A)-3', ... | | Authors: | Biswas, T, Aihara, H, Radman-Livaja, M, Filman, D, Landy, A, Ellenberger, T. | | Deposit date: | 2005-03-03 | | Release date: | 2005-06-28 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A structural basis for allosteric control of DNA recombination by lambda integrase.
Nature, 435, 2005
|
|
7Q1N
 
 | | Crystal structure of human butyrylcholinesterase in complex with N-[(2R)-3-[(cyclohexylmethyl)amino]-2-hydroxypropyl]-2,2-diphenylacetamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Brazzolotto, X, Panek, D, Pasieka, A, Malawska, B, Nachon, F. | | Deposit date: | 2021-10-20 | | Release date: | 2022-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of new, highly potent and selective inhibitors of BuChE - design, synthesis, in vitro and in vivo evaluation and crystallography studies.
Eur.J.Med.Chem., 249, 2023
|
|
5NK2
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2b | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-[[(3~{R},4~{R})-3-fluoranylpiperidin-4-yl]carbamoyl]phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
5NJ6
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in ternary complex with Fab3949 and AZ7188 at 4.0 angstrom resolution | | Descriptor: | Fab3949 H, Fab3949 L, Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2 | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
5NJZ
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1g | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-(piperidin-4-ylcarbamoyl)phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
6TE2
 
 | | Crystal structure of human protein kinase CK2alpha' (CSNK2A2 gene product) in complex with the 2-aminothiazole-type inhibitor 17 | | Descriptor: | 3-[(4-pyridin-2-yl-1,3-thiazol-2-yl)amino]benzoic acid, Casein kinase II subunit alpha' | | Authors: | Niefind, K, Lindenblatt, D, Jose, J, Applegate, V.M, Nickelsen, A. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.922 Å) | | Cite: | Structural and Mechanistic Basis of the Inhibitory Potency of Selected 2-Aminothiazole Compounds on Protein Kinase CK2.
J.Med.Chem., 63, 2020
|
|
5NKA
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2g | | Descriptor: | 1,2-ETHANEDIOL, 4-[[3-[[5-[(2-chloranyl-6-methyl-phenyl)carbamoyl]-1,3-thiazol-2-yl]amino]phenyl]carbonylamino]cyclohexane-1-carboxylic acid, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.377 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
2XBR
 
 | | Raman crystallography of Hen White Egg Lysozyme - Low X-ray dose (0.2 MGy) | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Carpentier, P, Royant, A, Weik, M, Bourgeois, D. | | Deposit date: | 2010-04-14 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Raman Assisted Crystallography Reveals a Mechanism of X-Ray Induced Reversible Disulfide Radical Formation
Structure, 18, 2010
|
|
5NK1
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 1k | | Descriptor: | 2-[[3-[(4-azanyl-6-methyl-1,3,5-triazin-2-yl)carbamoyl]phenyl]amino]-~{N}-(2-chloranyl-6-methyl-phenyl)-1,3-thiazole-5-carboxamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
6TE8
 
 | | Neck of native GTA particle computed with C12 symmetry | | Descriptor: | Adaptor protein Rcc01688, Phage portal protein, HK97 family | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
5NK6
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with Compound 2d | | Descriptor: | Ephrin type-A receptor 2, ~{N}-(2-chloranyl-6-methyl-phenyl)-2-[[3-[(4-oxidanylcyclohexyl)carbamoyl]phenyl]amino]-1,3-thiazole-5-carboxamide | | Authors: | Kudlinzki, D, Linhard, V.L, Witt, K, Gande, S.L, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.267 Å) | | Cite: | Chemoproteomics-Aided Medicinal Chemistry for the Discovery of EPHA2 Inhibitors.
ChemMedChem, 12, 2017
|
|
6TEB
 
 | | Tail-baseplate interface of native GTA particle computed with C6 symmetry | | Descriptor: | Distal tail protein Rcc01695, Tail tube protein Rcc01691 | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
2XCC
 
 | | Crystal structure of PcrH from Pseudomonas aeruginosa | | Descriptor: | REGULATORY PROTEIN PCRH | | Authors: | Job, V, Mattei, P.-J, Lemaire, D, Attree, I, Dessen, A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-05-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural Basis of Chaperone Recognition of Type III Secretion System Minor Translocator Proteins.
J.Biol.Chem., 285, 2010
|
|
6TFA
 
 | |
2XQH
 
 | | Crystal structure of an immunoglobulin-binding fragment of the trimeric autotransporter adhesin EibD | | Descriptor: | CHLORIDE ION, IMMUNOGLOBULIN-BINDING PROTEIN EIBD | | Authors: | Leo, J.C, Lyskowski, A, Hartmann, M, Schwarz, H, Linke, D, Lupas, A.N, Goldman, A. | | Deposit date: | 2010-09-02 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Structure of E. Coli Igg-Binding Protein D Suggests a General Model for Bending and Binding in Trimeric Autotransporter Adhesins.
Structure, 19, 2011
|
|
8DTM
 
 | | Cryo-EM structure of insulin receptor (IR) bound with S597 component 2 | | Descriptor: | Insulin mimetic peptide S597 component 2, Insulin receptor | | Authors: | Park, J, Li, J, Mayer, J.P, Ball, K.A, Wu, J.Y, Hall, C, Accili, D, Stowell, M.H.B, Bai, X.C, Choi, E. | | Deposit date: | 2022-07-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Activation of the insulin receptor by an insulin mimetic peptide.
Nat Commun, 13, 2022
|
|
2XFN
 
 | | Human monoamine oxidase B in complex with 2-(2-benzofuranyl)-2- imidazoline | | Descriptor: | 2-(2-BENZOFURANYL)-2-IMIDAZOLINE, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Bonivento, D, Milczek, E.M, McDonald, G.R, Binda, C, Holt, A, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2010-05-26 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potentiation of ligand binding through cooperative effects in monoamine oxidase B.
J. Biol. Chem., 285, 2010
|
|
8DPR
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with inhibitor TKB-248 | | Descriptor: | 2,2,2-trifluoro-N-{(2S)-1-[(1R,2S,5S)-2-({(2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamothioyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexan-3-yl]-3,3-dimethyl-1-oxobutan-2-yl}acetamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-16 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8DOY
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-198 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3C-like proteinase nsp5, ... | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-14 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Potent and biostable inhibitors of the main protease of SARS-CoV-2.
Iscience, 25, 2022
|
|
2XHW
 
 | | HCV-J4 NS5B Polymerase Trigonal Crystal Form | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
2XCG
 
 | | Tranylcypromine-inhibited human monoamine oxidase B in complex with 2- (2-benzofuranyl)-2-imidazoline | | Descriptor: | 2-(2-BENZOFURANYL)-2-IMIDAZOLINE, 3-PHENYLPROPANAL, Amine oxidase [flavin-containing] B, ... | | Authors: | Bonivento, D, Milczek, E.M, McDonald, G.R, Binda, C, Holt, A, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2010-04-23 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potentiation of ligand binding through cooperative effects in monoamine oxidase B.
J. Biol. Chem., 285, 2010
|
|
6TL0
 
 | | Solution structure and 1H, 13C and 15N chemical shift assignments for the complex of VPS29 with VARP 687-747 | | Descriptor: | Ankyrin repeat domain-containing protein 27, Vacuolar protein sorting-associated protein 29, ZINC ION | | Authors: | Owen, D.J, Neuhaus, D, Yang, J.-C, Crawley-Snowdon, H. | | Deposit date: | 2019-11-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanism and evolution of the Zn-fingernail required for interaction of VARP with VPS29.
Nat Commun, 11, 2020
|
|
8DTL
 
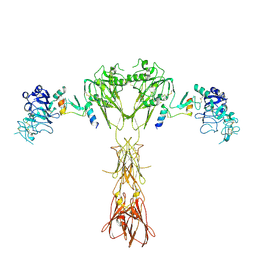 | | Cryo-EM structure of insulin receptor (IR) bound with S597 peptide | | Descriptor: | Insulin mimetic peptide S597, Insulin receptor | | Authors: | Park, J, Li, J, Mayer, J.P, Ball, K.A, Wu, J.Y, Hall, C, Accili, D, Stowell, M.H.B, Bai, X.C, Choi, E. | | Deposit date: | 2022-07-25 | | Release date: | 2022-09-07 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Activation of the insulin receptor by an insulin mimetic peptide.
Nat Commun, 13, 2022
|
|
