6I8E
 
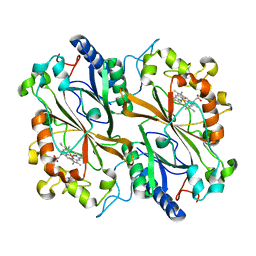 | | Dye type peroxidase Aa from Streptomyces lividans: 65.6 kGy structure | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | Deposit date: | 2018-11-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
6I91
 
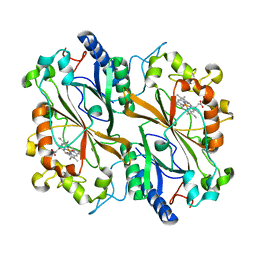 | | Dye type peroxidase Aa from Streptomyces lividans: 156.8 kGy structure | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ebrahim, A, Moreno-Chicano, T, Worrall, J.A.R, Strange, R.W, Axford, D, Sherrell, D.A, Appleby, M, Owen, R.L. | | Deposit date: | 2018-11-22 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dose-resolved serial synchrotron and XFEL structures of radiation sensitive metalloproteins
To be published
|
|
6H03
 
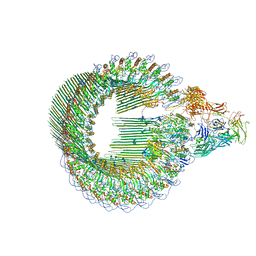 | | OPEN CONFORMATION OF THE MEMBRANE ATTACK COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5,Complement C5, ... | | Authors: | Menny, A, Serna, M, Boyd, C.M, Gardner, S, Joseph, A.P, Topf, M, Bubeck, D. | | Deposit date: | 2018-07-06 | | Release date: | 2018-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | CryoEM reveals how the complement membrane attack complex ruptures lipid bilayers.
Nat Commun, 9, 2018
|
|
6H26
 
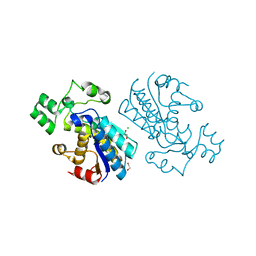 | | Rabbit muscle phosphoglycerate mutase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Phosphoglycerate mutase | | Authors: | Wisniewski, J, Barciszewski, J, Jaskolski, M, Rakus, D. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Rabbit muscle phosphoglycerate mutase
To Be Published
|
|
1MGY
 
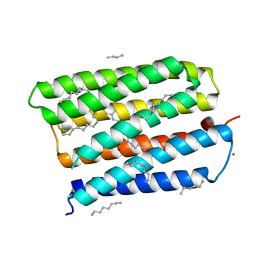 | | Structure of the D85S mutant of bacteriorhodopsin with bromide bound | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, BROMIDE ION, Bacteriorhodopsin, ... | | Authors: | Facciotti, M.T, Cheung, V.S, Nguyen, D, Rouhani, S, Glaeser, R.M. | | Deposit date: | 2002-08-16 | | Release date: | 2003-07-07 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Bromide-Bound D85S Mutant of Bacteriorhodopsin:
Principles of Ion Pumping
Biophys.J., 85, 2003
|
|
6BXH
 
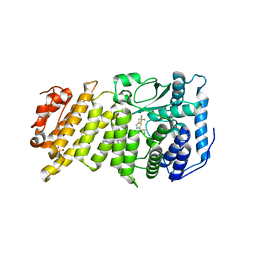 | | Menin in complex with MI-853 | | Descriptor: | 1-{2-[4-(fluoroacetyl)piperazin-1-yl]ethyl}-4-methyl-5-[(4-{[6-(2,2,2-trifluoroethyl)thieno[2,3-d]pyrimidin-4-yl]amino}piperidin-1-yl)methyl]-1H-indole-2-carbonitrile, DIMETHYL SULFOXIDE, Menin, ... | | Authors: | Borkin, D, Klossowski, S, Pollock, J, Linhares, B, Cierpicki, T, Grembecka, J. | | Deposit date: | 2017-12-18 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Menin in complex with MI-853
To Be Published
|
|
1MIX
 
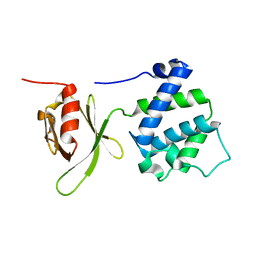 | | Crystal structure of a FERM domain of Talin | | Descriptor: | Talin | | Authors: | Garcia-Alvarez, B, de Pereda, J.M, Calderwood, D.A, Ulmer, T.S, Critchley, D, Campbell, I.D, Ginsberg, M.H, Liddington, R.C. | | Deposit date: | 2002-08-23 | | Release date: | 2003-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural determinants of integrin recognition by talin
Mol.Cell, 11, 2003
|
|
4CK0
 
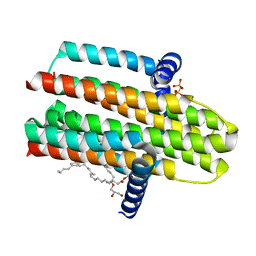 | | Crystal structure of the integral membrane diacylglycerol kinase - form 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DIACYLGLYCEROL KINASE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Li, D, Lyons, J.A, Vogeley, L, Aragao, D, Caffrey, M. | | Deposit date: | 2013-12-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.924 Å) | | Cite: | Crystal Structure of the Integral Membrane Diacylglycerol Kinase with Zn-Amppcp Bound and its Catalytic Mechanism
To be Published
|
|
1MIZ
 
 | | Crystal structure of an integrin beta3-talin chimera | | Descriptor: | TALIN, integrin beta3 | | Authors: | Garcia-Alvarez, B, de Pereda, J.M, Calderwood, D.A, Ulmer, T.S, Critchley, D, Campbell, I.D, Ginsberg, M.H, Liddington, R.C. | | Deposit date: | 2002-08-23 | | Release date: | 2003-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants of integrin recognition by talin
Mol.Cell, 11, 2003
|
|
4CJZ
 
 | | Crystal structure of the integral membrane diacylglycerol kinase DgkA- 9.9, delta 4 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DIACYLGLYCEROL KINASE | | Authors: | Li, D, Aragao, D, Pye, V.E, Caffrey, M. | | Deposit date: | 2013-12-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal Structure of the Integral Membrane Diacylglycerol Kinase with Zn-Amppcp Bound and its Catalytic Mechanism
To be Published
|
|
6BXX
 
 | | GYNGFG from low-complexity domain of hnRNPA1, residues 243-248 | | Descriptor: | hnRNPA1 | | Authors: | Hughes, M.P, Rodriguez, J.A, Sawaya, M.R, Cascio, D, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2017-12-19 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic structures of low-complexity protein segments reveal kinked beta sheets that assemble networks.
Science, 359, 2018
|
|
6ZPA
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dA and one catalytic Zn2+ ion | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, DatZ, LITHIUM ION, ... | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.86000258 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
6ZPC
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DatZ, LITHIUM ION, ... | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.2683593 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
6C1E
 
 | | NavAb NormoPP mutant | | Descriptor: | (3ALPHA,5ALPHA,7ALPHA,8ALPHA,12ALPHA,14BETA,17ALPHA)-3,7,12-TRIHYDROXYCHOL-1-EN-24-AMIDE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 5-BETA-24-NOR-CHOLANE-3(ALPHA),7(ALPHA),12(ALPHA)-TRIOL, ... | | Authors: | Catterall, W.A, Zheng, N, Jiang, D, Gamal El-Din, T.M. | | Deposit date: | 2018-01-04 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for gating pore current in periodic paralysis.
Nature, 557, 2018
|
|
6ZP9
 
 | |
6ZPB
 
 | | Cyanophage S-2L HD phosphohydrolase (DatZ) bound to dA and two catalytic Co2+ ions | | Descriptor: | (2R,3S,5R)-5-(6-amino-9H-purin-9-yl)-tetrahydro-2-(hydroxymethyl)furan-3-ol, COBALT (II) ION, DatZ | | Authors: | Czernecki, D, Legrand, P, Delarue, M. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.72097385 Å) | | Cite: | How cyanophage S-2L rejects adenine and incorporates 2-aminoadenine to saturate hydrogen bonding in its DNA.
Nat Commun, 12, 2021
|
|
7A9A
 
 | | Crystal structure of rubredoxin B (Rv3250c) from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vakhrameev, D, Kavaleuski, A, Bukhdruker, S, Marin, E, Sushko, T, Grabovec, I.P, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2020-09-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | A new twist of rubredoxin function in M. tuberculosis.
Bioorg.Chem., 109, 2021
|
|
1MN7
 
 | | NDP kinase mutant (H122G;N119S;F64W) in complex with aBAZTTP | | Descriptor: | 3'-AZIDO-3'-DEOXY-THYMIDINE-5'-ALPHA BORANO TRIPHOSPHATE, MAGNESIUM ION, NDP kinase | | Authors: | gallois-montbrun, s, schneider, b, chen, y, giacomoni-fernandes, v, mulard, l, morera, s, janin, j, deville-bonne, d, veron, m. | | Deposit date: | 2002-09-05 | | Release date: | 2002-10-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Improving nucleoside diphosphate kinase for antiviral nucleotide analogs activation
J.BIOL.CHEM., 277, 2002
|
|
6H1S
 
 | | Structure of the BM3 heme domain in complex with fluconazole | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Jeffreys, L.N, Munro, A.W.M, Leys, D. | | Deposit date: | 2018-07-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel insights into P450 BM3 interactions with FDA-approved antifungal azole drugs.
Sci Rep, 9, 2019
|
|
4ONG
 
 | | Fab fragment of 3D6 in complex with amyloid beta 1-40 | | Descriptor: | 3D6 FAB ANTIBODY HEAVY CHAIN, 3D6 FAB ANTIBODY LIGHT CHAIN, Amyloid beta A4 protein, ... | | Authors: | Feinberg, H, Saldanha, J.W, Diep, L, Goel, A, Widom, A, Veldman, G.M, Weis, W.I, Schenk, D, Basi, G.S. | | Deposit date: | 2014-01-28 | | Release date: | 2014-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure reveals conservation of amyloid-beta conformation recognized by 3D6 following humanization to bapineuzumab.
Alzheimers Res Ther, 6, 2014
|
|
4HEP
 
 | | Complex of lactococcal phage TP901-1 with a llama vHH (vHH17) binder (nanobody) | | Descriptor: | BPP, SULFATE ION, vHH17 domain | | Authors: | Desmyter, A, Spinelli, S, Farenc, C, Blangy, S, Bebeacua, C, van Sinderen, D, Mahony, J, Cambillau, C. | | Deposit date: | 2012-10-04 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Viral infection modulation and neutralization by camelid nanobodies
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6H5Q
 
 | | Cryo-EM structure of in vitro assembled Measles virus N into nucleocapsid-like particles (NCLPs) bound to polyA RNA hexamers. | | Descriptor: | Nucleocapsid, RNA (5'-R(*AP*AP*AP*AP*AP*A)-3') | | Authors: | Desfosses, A, Milles, S, Ringkjobing Jensen, M, Guseva, S, Colletier, J, Maurin, D, Schoehn, G, Gutsche, I, Ruigrok, R, Blackledge, M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4HEM
 
 | | Llama vHH-02 binder of ORF49 (RBP) from lactococcal phage TP901-1 | | Descriptor: | Anti-baseplate TP901-1 Llama vHH 02, BPP | | Authors: | Desmyter, A, Spinelli, S, Farenc, C, Blangy, S, Bebeacua, C, van Sinderen, D, Mahony, J, Cambillau, C. | | Deposit date: | 2012-10-04 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Viral infection modulation and neutralization by camelid nanobodies
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1MQP
 
 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-6 | | Descriptor: | GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Geobacillus stearothermophilus alpha-glucuronidase complexed with its substrate and products: mechanistic implications.
J.Biol.Chem., 279, 2004
|
|
6GZS
 
 | |
