7QPW
 
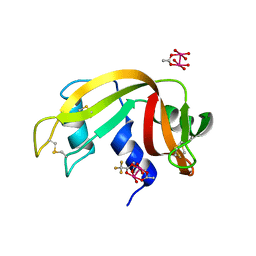 | |
7QCJ
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2,4-dihydroxybenzylidene)-thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(2,4-dihydroxybenzylidene)-thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
7QPY
 
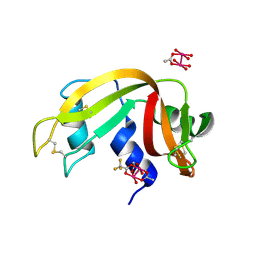 | |
7QCH
 
 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(3,5-dimethoxy-4-hydroxybenzyliden)thiosemicarbazone | | Descriptor: | CHLORIDE ION, GLYCEROL, N-(3,5-dimetoxy-4-hydroxybenzyliden)thiosemicarbazone, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
1W5O
 
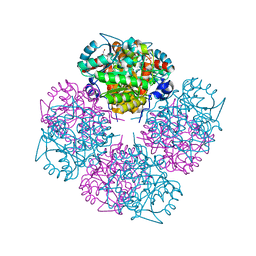 | | Stepwise introduction of zinc binding site into porphobilinogen synthase of Pseudomonas aeruginosa (mutations A129C, D131C and D139C) | | Descriptor: | DELTA-AMINOLEVULINIC ACID DEHYDRATASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Frere, F, Reents, H, Schubert, W.-D, Heinz, D.W, Jahn, D. | | Deposit date: | 2004-08-09 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tracking the Evolution of Porphobilinogen Synthase Metal Dependence in Vitro
J.Mol.Biol., 345, 2005
|
|
7QCG
 
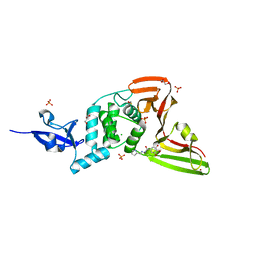 | | Structure of SARS-CoV-2 Papain-like Protease bound to N-(2-pyrrolidyl)-3,4,5-trihydroxybenzoylhydrazone | | Descriptor: | 3,4,5-tris(oxidanyl)-N-[(E)-1H-pyrrol-2-ylmethylideneamino]benzamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ewert, W, Gunther, S, Reinke, P, Falke, S, Lieske, J, Miglioli, F, Carcelli, M, Srinivasan, V, Betzel, C, Han, H, Lorenzen, K, Guenther, C, Niebling, S, Garcia-Alai, M, Hinrichs, W, Rogolino, D, Meents, A. | | Deposit date: | 2021-11-23 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hydrazones and Thiosemicarbazones Targeting Protein-Protein-Interactions of SARS-CoV-2 Papain-like Protease.
Front Chem, 10, 2022
|
|
1W4L
 
 | | Complex of TcAChE with bis-acting galanthamine derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Greenblatt, H.M, Guillou, C, Guenard, D, Badet, B, Thal, C, Silman, I, Sussman, J.L. | | Deposit date: | 2004-07-25 | | Release date: | 2004-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The complex of a bivalent derivative of galanthamine with torpedo acetylcholinesterase displays drastic deformation of the active-site gorge: implications for structure-based drug design.
J.Am.Chem.Soc., 126, 2004
|
|
1W6R
 
 | | Complex of TcAChE with galanthamine derivative | | Descriptor: | (-)-GALANTHAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Greenblatt, H.M, Guillou, C, Guenard, D, Badet, B, Thal, C, Silman, I, Sussman, J.L. | | Deposit date: | 2004-08-23 | | Release date: | 2004-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The complex of a bivalent derivative of galanthamine with torpedo acetylcholinesterase displays drastic deformation of the active-site gorge: implications for structure-based drug design.
J.Am.Chem.Soc., 126, 2004
|
|
6J4T
 
 | | Crystal structure of arabidopsis ADAL complexed with IMP | | Descriptor: | Adenosine/AMP deaminase family protein, INOSINIC ACID, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
1W4Y
 
 | | Ferrous horseradish peroxidase C1A in complex with carbon monoxide | | Descriptor: | CALCIUM ION, CARBON MONOXIDE, HORSERADISH PEROXIDASE C1A, ... | | Authors: | Carlsson, G.H, Nicholls, P, Svistunenko, D, Berglund, G.I, Hajdu, J. | | Deposit date: | 2004-08-03 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Complexes of Horseradish Peroxidase with Formate, Acetate, and Carbon Monoxide
Biochemistry, 44, 2005
|
|
6J14
 
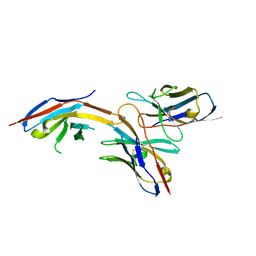 | | Complex structure of GY-14 and PD-1 | | Descriptor: | GY-14 heavy chain V fragment, GY-14 light chain V fragment, Programmed cell death protein 1 | | Authors: | Chen, D, Tan, S, Whang, H, Zhang, H, Chai, Y, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2018-12-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The FG Loop of PD-1 Serves as a "Hotspot" for Therapeutic Monoclonal Antibodies in Tumor Immune Checkpoint Therapy.
Iscience, 14, 2019
|
|
1JTP
 
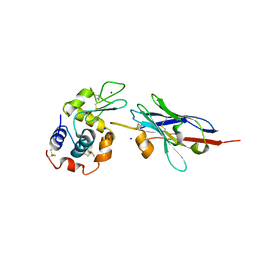 | | Degenerate interfaces in antigen-antibody complexes | | Descriptor: | FORMIC ACID, LYSOZYME C, SODIUM ION, ... | | Authors: | Decanniere, K, Transue, T.R, Desmyter, A, Maes, D, Muyldermans, S, Wyns, L. | | Deposit date: | 2001-08-21 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Degenerate interfaces in antigen-antibody complexes.
J.Mol.Biol., 313, 2001
|
|
1JUJ
 
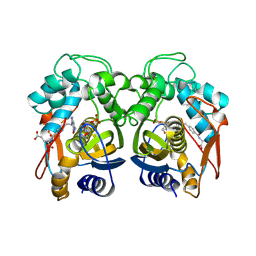 | | Human Thymidylate Synthase Bound to dUMP and LY231514, a Pyrrolo(2,3-d)pyrimidine-based Antifolate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Sayre, P.H, Finer-Moore, J.S, Fritz, T.A, Biermann, D, Gates, S.B, MacKellar, W.C, Patel, V.F, Stroud, R.M. | | Deposit date: | 2001-08-24 | | Release date: | 2001-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases.
J.Mol.Biol., 313, 2001
|
|
1WAO
 
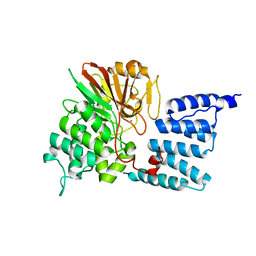 | | PP5 structure | | Descriptor: | MANGANESE (II) ION, SERINE/THREONINE PROTEIN PHOSPHATASE 5 | | Authors: | Barford, D. | | Deposit date: | 2004-10-27 | | Release date: | 2005-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for Tpr Domain-Mediated Regulation of Protein Phosphatase 5
Embo J., 24, 2005
|
|
1W9D
 
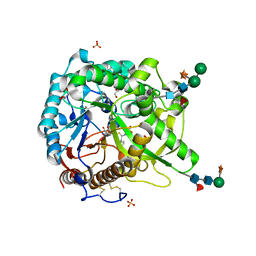 | | S. alba myrosinase in complex with S-ethyl phenylacetothiohydroximate- O-sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Bourderioux, A, Lefoix, M, Gueyrard, D, Tatibouet, A, Cottaz, S, Arzt, S, Burmeister, W.P, Rollin, P. | | Deposit date: | 2004-10-08 | | Release date: | 2005-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Glucosinolate-Myrosinase System. New Insights Into Enzyme-Substrate Interactions by Use of Simplified Inhibitors
Org.Biomol.Chem., 3, 2005
|
|
6J81
 
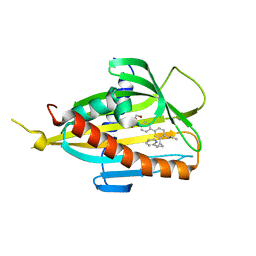 | | Crystal structure of CERT START domain in complex with compound E14 | | Descriptor: | 2-[4-[4-propyl-3-[(1~{S},2~{R})-2-pyridin-2-ylcyclopropyl]phenyl]phenyl]sulfonylethanol, LIPID-TRANSFER PROTEIN CERT | | Authors: | Suzuki, M, Nakao, N, Ueno, M, Sakai, S, Egawa, D, Hanzawa, H, Kawasaki, S, Kumagai, K, Kobayashi, S, Hanada, K. | | Deposit date: | 2019-01-18 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Natural ligand-nonmimetic inhibitors of the lipid-transfer protein CERT
Commun Chem, 2019
|
|
5JP1
 
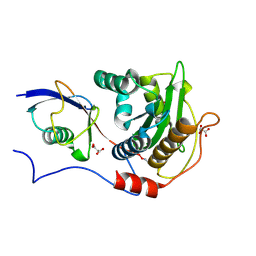 | | Structure of Xanthomonas campestris effector protein XopD bound to tomato SUMO | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, MALONATE ION, Small ubiquitin-related modifier, ... | | Authors: | Pruneda, J.N, Komander, D. | | Deposit date: | 2016-05-03 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Molecular Basis for Ubiquitin and Ubiquitin-like Specificities in Bacterial Effector Proteases.
Mol.Cell, 63, 2016
|
|
1W5Y
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,5-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXANEDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
6J90
 
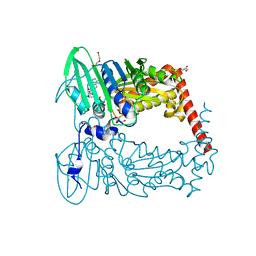 | | Crystal Structure of GyraseB N-Terminal Domain complex with ATP from Salmonella Typhi at 2.2A Resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kaur, G, Sachdeva, E, Tiwari, P, Gupta, D, Ethayathulla, A.S, Kaur, P. | | Deposit date: | 2019-01-21 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of GyraseB N-Terminal Domain complex with ATP from Salmonella Typhi at 2.2A Resolution
To Be Published
|
|
5JPQ
 
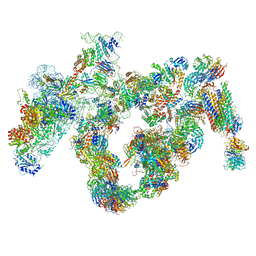 | | Cryo-EM structure of the 90S pre-ribosome | | Descriptor: | 18S ribosomal RNA, Bms1, Emg1, ... | | Authors: | Turk, M, Cheng, J, Berninghausen, O, Kornprobst, M, Flemming, D, Kos-Braun, I.C, Kos, M, Thoms, M, Hurt, E, Beckmann, R. | | Deposit date: | 2016-05-04 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Architecture of the 90S Pre-ribosome: A Structural View on the Birth of the Eukaryotic Ribosome.
Cell, 166, 2016
|
|
1W8T
 
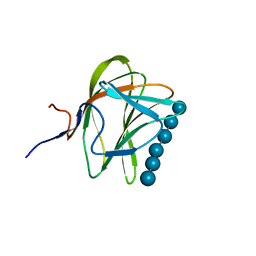 | | CBM29-2 mutant K74A complexed with cellulohexaose: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-28 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
5KBD
 
 | |
1VYQ
 
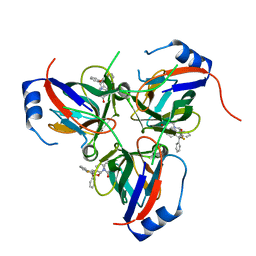 | | Novel inhibitors of Plasmodium Falciparum dUTPase provide a platform for anti-malarial drug design | | Descriptor: | 2,3-DEOXY-3-FLUORO-5-O-TRITYLURIDINE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Whittingham, J.L, Leal, I, Kasinathan, G, Nguyen, C, Bell, E, Jones, A.F, Berry, C, Benito, A, Turkenburg, J.P, Dodson, E.J, Ruiz Perez, L.M, Wilkinson, A.J, Johansson, N.G, Brun, R, Gilbert, I.H, Gonzalez Pacanowska, D, Wilson, K.S. | | Deposit date: | 2004-05-05 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dutpase as a Platform for Antimalarial Drug Design: Structural Basis for the Selectivity of a Class of Nucleoside Inhibitors.
Structure, 13, 2005
|
|
5JWE
 
 | | Crystal structure of H-2Db in complex with the LCMV-derived GP92-101 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Buratto, J, Badia-Martinez, D, Norstrom, M, Sandalova, T, Achour, A. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of H-2Db in complex with the LCMV-derived peptides GP92 and GP392 explain pleiotropic effects of glycosylation on antigen presentation and immunogenicity.
PLoS ONE, 12, 2017
|
|
1VRZ
 
 | | Helix turn helix motif | | Descriptor: | ACETATE ION, DE NOVO DESIGNED 21 RESIDUE PEPTIDE | | Authors: | Rudresh, Ramakumar, S, Ramagopal, U.A, Inai, Y, Sahal, D. | | Deposit date: | 2005-10-14 | | Release date: | 2005-11-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | De Novo Design and Characterization of a Helical Hairpin Eicosapeptide; Emergence of an Anion Receptor in the Linker Region.
Structure, 12, 2004
|
|
