1OCE
 
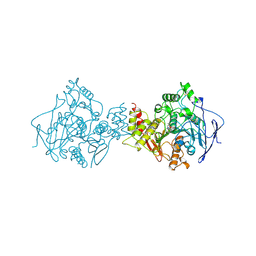 | | ACETYLCHOLINESTERASE (E.C. 3.1.1.7) COMPLEXED WITH MF268 | | Descriptor: | ACETYLCHOLINESTERASE, CIS-2,6-DIMETHYLMORPHOLINOOCTYLCARBAMYLESEROLINE | | Authors: | Bartolucci, C, Perola, E, Cellai, L, Brufani, M, Lamba, D. | | Deposit date: | 1998-06-12 | | Release date: | 1999-05-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | "Back door" opening implied by the crystal structure of a carbamoylated acetylcholinesterase.
Biochemistry, 38, 1999
|
|
6N3J
 
 | |
6RWB
 
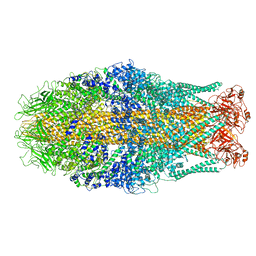 | | Cryo-EM structure of Yersinia pseudotuberculosis TcaA-TcaB | | Descriptor: | Toxin,Toxin complex subunit TcaB,Putative toxin subunit,Putative toxin subunit,Toxin,Toxin complex subunit TcaB,Putative toxin subunit,Putative toxin subunit,Toxin,Toxin complex subunit TcaB,Putative toxin subunit,Putative toxin subunit,Toxin,Toxin complex subunit TcaB,Putative toxin subunit,Putative toxin subunit | | Authors: | Roderer, D, Leidreiter, F, Gatsogiannis, C, Meusch, D, Benz, R, Raunser, S. | | Deposit date: | 2019-06-04 | | Release date: | 2019-10-23 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Common architecture of Tc toxins from human and insect pathogenic bacteria.
Sci Adv, 5, 2019
|
|
4ET9
 
 | | Hen egg-white lysozyme solved from 5 fs free-electron laser pulse data | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
4ETD
 
 | | Lysozyme, room-temperature, rotating anode, 0.0026 MGy | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
2PZY
 
 | | Structure of MK2 Complexed with Compound 76 | | Descriptor: | (4R)-N-[4-({[2-(DIMETHYLAMINO)ETHYL]AMINO}CARBONYL)-1,3-THIAZOL-2-YL]-4-METHYL-1-OXO-2,3,4,9-TETRAHYDRO-1H-BETA-CARBOLINE-6-CARBOXAMIDE, MAP kinase-activated protein kinase 2, STAUROSPORINE | | Authors: | White, A, Wu, J.P, Wang, J, Abeywardane, A, Andersen, D, Emmanuel, M, Gautschi, E, Goldberg, D.R, Kashem, M.A, Lukas, S, Mao, W, Martin, L, Morwick, T, Moss, N, Pargellis, C, Patel, U.R, Patnaude, L, Peet, G.W, Skow, D, Snow, R.J, Ward, Y, Werneburg, B. | | Deposit date: | 2007-05-18 | | Release date: | 2007-07-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The discovery of carboline analogs as potent MAPKAP-K2 inhibitors
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2PY1
 
 | |
2PX7
 
 | | Crystal structure of 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Thermus thermophilus HB8 | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | | Authors: | Chen, L, Tsukuda, M, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Chen, L.-Q, Liu, Z.-J, Lee, D, Chang, S.-H, Nguyen, D, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-14 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase from Thermus thermophilus HB8.
To be Published
|
|
2P64
 
 | | D domain of b-TrCP | | Descriptor: | CADMIUM ION, F-box/WD repeat protein 1A | | Authors: | Neculai, D, Orlicky, S, Ceccarelli, D. | | Deposit date: | 2007-03-16 | | Release date: | 2007-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination.
Cell(Cambridge,Mass.), 129, 2007
|
|
2FH7
 
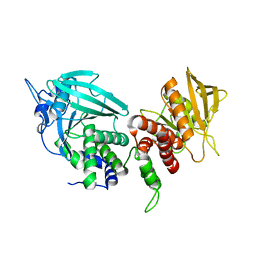 | | Crystal structure of the phosphatase domains of human PTP SIGMA | | Descriptor: | Receptor-type tyrosine-protein phosphatase S | | Authors: | Alvarado, J, Udupi, R, Smith, D, Koss, J, Wasserman, S.R, Ozyurt, S, Atwell, S, Powell, A, Kearins, M.C, Rooney, I, Maletic, M, Bain, K.T, Freeman, J.C, Russell, M, Thompson, D.A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-12-23 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
3GIG
 
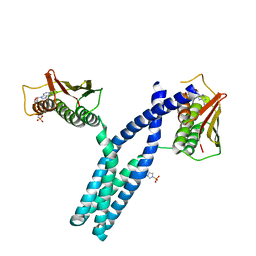 | | Crystal structure of phosphorylated DesKC in complex with AMP-PCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Sensor histidine kinase desK | | Authors: | Trajtenberg, F, Albanesi, D, Alzari, P.M, Buschiazzo, A, de Mendoza, D. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Structural plasticity and catalysis regulation of a thermosensor histidine kinase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2EXV
 
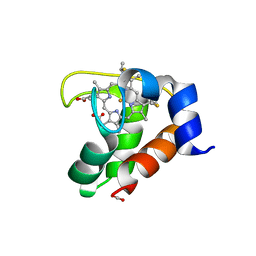 | | Crystal structure of the F7A mutant of the cytochrome c551 from Pseudomonas aeruginosa | | Descriptor: | ACETIC ACID, Cytochrome c-551, HEME C | | Authors: | Bonivento, D, Di Matteo, A, Borgia, A, Travaglini-Allocatelli, C, Brunori, M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-02-07 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Unveiling a Hidden Folding Intermediate in c-Type Cytochromes by Protein Engineering
J.Biol.Chem., 281, 2006
|
|
1O6Z
 
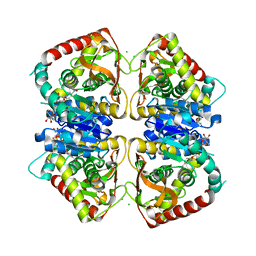 | | 1.95 A resolution structure of (R207S,R292S) mutant of malate dehydrogenase from the halophilic archaeon Haloarcula marismortui (holo form) | | Descriptor: | CHLORIDE ION, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Irimia, A, Ebel, C, Madern, D, Richard, S.B, Cosenza, L.W, Zaccai, G, Vellieux, F.M.D. | | Deposit date: | 2002-10-22 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Oligomeric States of Haloarcula Marismortui Malate Dehydrogenase are Modulated by Solvent Components as Shown by Crystallographic and Biochemical Studies
J.Mol.Biol., 326, 2003
|
|
2Q9M
 
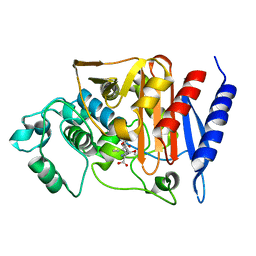 | | 4-Substituted Trinems as Broad Spectrum-Lactamase Inhibitors: Structure-based Design, Synthesis and Biological Activity | | Descriptor: | (1R,4S,7AS)-1-(1-FORMYLPROP-1-EN-1-YL)-4-METHOXY-2,4,5,6,7,7A-HEXAHYDRO-1H-ISOINDOLE-3-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Plantan, I, Selic, L, Mesar, T, Stefanic Anderluh, P, Oblak, M, Prezelj, A, Hesse, L, Andrejasic, M, Vilar, M, Turk, D, Kocijan, A, Prevec, T, Vilfan, G, Kocjan, D, Copar, A, Urleb, U, Solmajer, T. | | Deposit date: | 2007-06-13 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 4-Substituted Trinems as Broad Spectrum beta-Lactamase Inhibitors: Structure-Based Design, Synthesis, and Biological Activity
J.Med.Chem., 50, 2007
|
|
4ETA
 
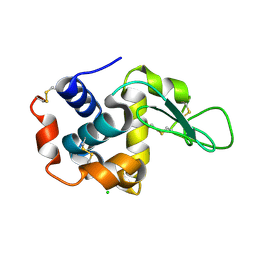 | | Lysozyme, room temperature, 400 kGy dose | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
3GIF
 
 | | Crystal structure of DesKC_H188E in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Sensor histidine kinase desK | | Authors: | Trajtenberg, F, Albanesi, D, Alzari, P.M, Buschiazzo, A, de Mendoza, D. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity and catalysis regulation of a thermosensor histidine kinase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4ET8
 
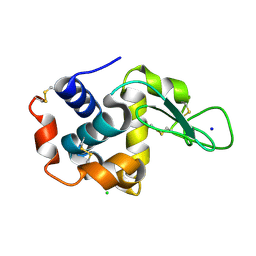 | | Hen egg-white lysozyme solved from 40 fs free-electron laser pulse data | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Boutet, S, Lomb, L, Williams, G, Barends, T, Aquila, A, Doak, R.B, Weierstall, U, DePonte, D, Steinbrener, J, Shoeman, R, Messerschmidt, M, Barty, A, White, T, Kassemeyer, S, Kirian, R, Seibert, M, Montanez, P, Kenney, C, Herbst, R, Hart, P, Pines, J, Haller, G, Gruner, S, Philllip, H, Tate, M, Hromalik, M, Koerner, L, van Bakel, N, Morse, J, Ghonsalves, W, Arnlund, D, Bogan, M, Calemann, C, Fromme, R, Hampton, C, Hunter, M, Johansson, L, Katona, G, Kupitz, C, Liang, M, Martin, A, Nass, K, Redecke, L, Stellato, F, Timneanu, N, Wang, D, Zatsepin, N, Schafer, D, Defever, K, Neutze, R, Fromme, P, Spence, J, Chapman, H, Schlichting, I. | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution protein structure determination by serial femtosecond crystallography.
Science, 337, 2012
|
|
3GIE
 
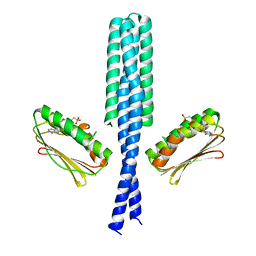 | | Crystal structure of DesKC_H188E in complex with AMP-PCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Sensor histidine kinase desK | | Authors: | Trajtenberg, F, Albanesi, D, Alzari, P.M, Buschiazzo, A, de Mendoza, D. | | Deposit date: | 2009-03-05 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural plasticity and catalysis regulation of a thermosensor histidine kinase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2DRP
 
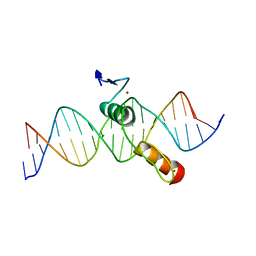 | | THE CRYSTAL STRUCTURE OF A TWO ZINC-FINGER PEPTIDE REVEALS AN EXTENSION TO THE RULES FOR ZINC-FINGER/DNA RECOGNITION | | Descriptor: | DNA (5'-D(*CP*TP*AP*AP*TP*AP*AP*GP*GP*AP*TP*AP*AP*CP*GP*TP*C P*CP*G)-3'), DNA (5'-D(*TP*CP*GP*GP*AP*CP*GP*TP*TP*AP*TP*CP*CP*TP*TP*AP*T P*TP*A)-3'), PROTEIN (TRAMTRACK DNA-BINDING DOMAIN), ... | | Authors: | Fairall, L, Schwabe, J.W.R, Chapman, L, Finch, J.T, Rhodes, D. | | Deposit date: | 1994-06-06 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition.
Nature, 366, 1993
|
|
2Q7W
 
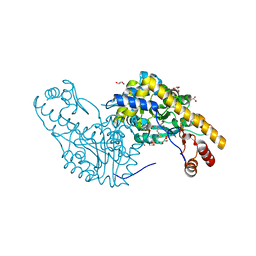 | | Structural Studies Reveals the Inactivation of E. coli L-aspartate aminotransferase (S)-4,5-amino-dihydro-2-thiophenecarboxylic acid (SADTA) via two mechanisms at pH 6.0 | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-07 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inactivation of Escherichia coli l-Aspartate Aminotransferase by (S)-4-Amino-4,5-dihydro-2-thiophenecarboxylic Acid Reveals "A Tale of Two Mechanisms".
Biochemistry, 46, 2007
|
|
2Q9N
 
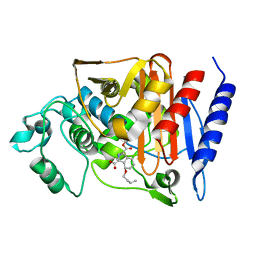 | | 4-Substituted Trinems as Broad Spectrum-Lactamase Inhibitors: Structure-based Design, Synthesis and Biological Activity | | Descriptor: | (1S,4R,7AR)-4-BUTOXY-1-[(1R)-1-FORMYLPROPYL]-2,4,5,6,7,7A-HEXAHYDRO-1H-ISOINDOLE-3-CARBOXYLIC ACID, Beta-lactamase | | Authors: | Plantan, I, Selic, L, Mesar, T, Stefanic Anderluh, P, Oblak, M, Prezelj, A, Hesse, L, Andrejasic, M, Vilar, M, Turk, D, Kocijan, A, Prevec, T, Vilfan, G, Kocjan, D, Copar, A, Urleb, U, Solmajer, T. | | Deposit date: | 2007-06-13 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 4-Substituted Trinems as Broad Spectrum beta-Lactamase Inhibitors: Structure-Based Design, Synthesis, and Biological Activity
J.Med.Chem., 50, 2007
|
|
2QB2
 
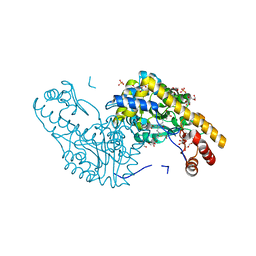 | | Structural Studies Reveal the Inactivation of E. coli L-aspartate aminotransferase by (s)-4,5-dihydro-2thiophenecarboylic acid (SADTA) via two mechanisms (at pH 7.0). | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]THIOPHENE-2-CARBOXYLIC ACID, Aspartate aminotransferase, ... | | Authors: | Liu, D, Pozharski, E, Lepore, B, Fu, M, Silverman, R.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-06-15 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inactivation of Escherichia coli L-aspartate aminotransferase by (S)-4-amino-4,5-dihydro-2-thiophenecarboxylic acid reveals "a tale of two mechanisms".
Biochemistry, 46, 2007
|
|
1QRZ
 
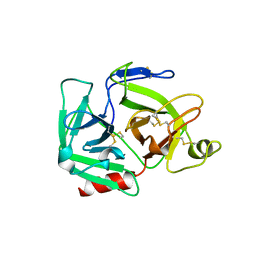 | | CATALYTIC DOMAIN OF PLASMINOGEN | | Descriptor: | PLASMINOGEN | | Authors: | Peisach, E, Wang, J, de los Santos, T, Reich, E, Ringe, D. | | Deposit date: | 1999-06-16 | | Release date: | 1999-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the proenzyme domain of plasminogen.
Biochemistry, 38, 1999
|
|
2D94
 
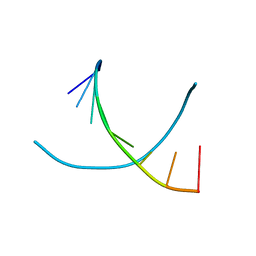 | | THE CONFORMATION OF THE DNA DOUBLE HELIX IN THE CRYSTAL IS DEPENDENT ON ITS ENVIRONMENT | | Descriptor: | DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*C)-3') | | Authors: | Shakked, Z, Guerstein-Guzikevich, G, Eisenstein, M, Frolow, F, Rabinovich, D. | | Deposit date: | 1993-07-13 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The conformation of the DNA double helix in the crystal is dependent on its environment.
Nature, 342, 1989
|
|
2D3T
 
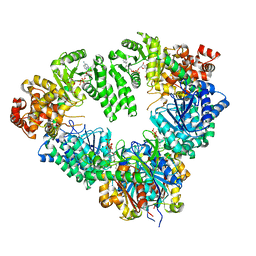 | | Fatty Acid beta-oxidation multienzyme complex from Pseudomonas Fragi, Form V | | Descriptor: | 3-ketoacyl-CoA thiolase, ACETYL COENZYME *A, Fatty oxidation complex alpha subunit, ... | | Authors: | Tsuchiya, D, Shimizu, N, Ishikawa, M, Suzuki, Y, Morikawa, K. | | Deposit date: | 2005-10-01 | | Release date: | 2006-02-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ligand-Induced Domain Rearrangement of Fatty Acid beta-Oxidation Multienzyme Complex
Structure, 14, 2006
|
|
