3ENI
 
 | | Crystal structure of the Fenna-Matthews-Olson Protein from Chlorobaculum Tepidum | | 分子名称: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll a protein | | 著者 | Tronrud, D, Camara-Artigas, A, Blankenship, R, Allen, J.P. | | 登録日 | 2008-09-25 | | 公開日 | 2009-05-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria.
Photosynth.Res., 100, 2009
|
|
2M8E
 
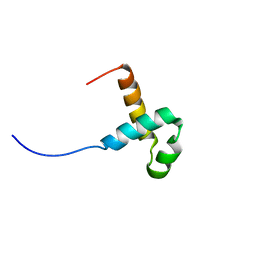 | | NMR structure of the PAI subdomain of Sleeping Beauty transposase | | 分子名称: | SLEEPING BEAUTY TRANSPOSASE | | 著者 | Eubanks, C, Schreifels, J, Aronovich, E, Carlson, D, Hacjkett, P, Nesmelova, I. | | 登録日 | 2013-05-17 | | 公開日 | 2013-12-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structural analysis of Sleeping Beauty transposase binding to DNA.
Protein Sci., 23, 2014
|
|
6S4A
 
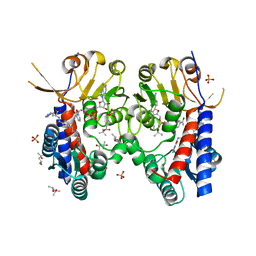 | | Structure of human MTHFD2 in complex with TH9028 | | 分子名称: | (2~{S})-2-[[5-[[2,4-bis(azanyl)-6-oxidanylidene-5~{H}-pyrimidin-5-yl]carbamoylamino]pyridin-2-yl]carbonylamino]-4-(1~{H}-1,2,3,4-tetrazol-5-yl)butanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | 著者 | Gustafsson, R, Scaletti, E.R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | 登録日 | 2019-06-27 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
6S4F
 
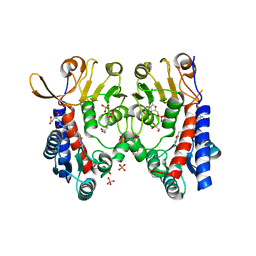 | | Structure of human MTHFD2 in complex with TH9619 | | 分子名称: | (E,4S)-4-[[5-[2-[2,6-bis(azanyl)-4-oxidanylidene-1H-pyrimidin-5-yl]ethanoylamino]-3-fluoranyl-pyridin-2-yl]carbonylamino]pent-2-enedioic acid, ADENOSINE-5'-DIPHOSPHATE, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | 著者 | Scaletti, E.R, Gustafsson, R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | 登録日 | 2019-06-27 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
6S4E
 
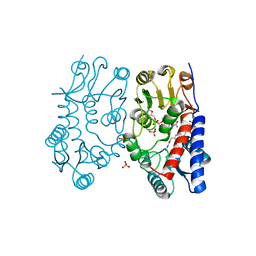 | | Structure of human MTHFD2 in complex with TH7299 | | 分子名称: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | 著者 | Gustafsson, R, Scaletti, E.R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | 登録日 | 2019-06-27 | | 公開日 | 2021-07-07 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
3ER3
 
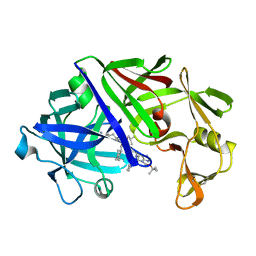 | | The active site of aspartic proteinases | | 分子名称: | 6-ammonio-N-[(2R,4R,5R)-5-{[N-(tert-butoxycarbonyl)-L-phenylalanyl-3-(1H-imidazol-3-ium-4-yl)-L-alanyl]amino}-6-cyclohexyl-4-hydroxy-2-(2-methylpropyl)hexanoyl]-L-norleucylphenylalanine, ENDOTHIAPEPSIN | | 著者 | Al-Karadaghi, S, Cooper, J.B, Veerapandian, B, Hoover, D, Blundell, T.L. | | 登録日 | 1991-01-02 | | 公開日 | 1991-04-15 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
1F0L
 
 | |
1F2H
 
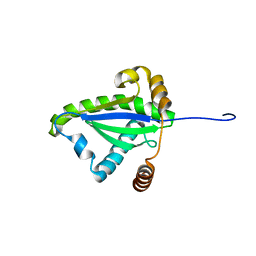 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF THE TNFR1 ASSOCIATED PROTEIN, TRADD. | | 分子名称: | TUMOR NECROSIS FACTOR RECEPTOR TYPE 1 ASSOCIATED DEATH DOMAIN PROTEIN | | 著者 | Tsao, D, McDonaugh, T, Malakian, K, Xu, G.-Y, Telliez, J.-B, Hsu, H, Lin, L.-L. | | 登録日 | 2000-05-24 | | 公開日 | 2001-05-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of N-TRADD and characterization of the interaction of N-TRADD and C-TRAF2, a key step in the TNFR1 signaling pathway.
Mol.Cell, 5, 2000
|
|
2MCY
 
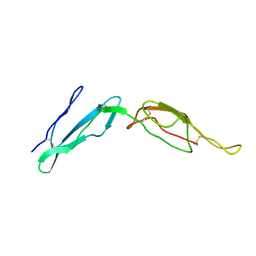 | | CR1 Sushi domains 2 and 3 | | 分子名称: | Complement receptor type 1 | | 著者 | Park, H.J, Guariento, M.J, Maciejewski, M, Hauart, R, Tham, W, Cowman, A.F, Schmidt, C.Q, Martens, H, Liszewski, K.M, Hourcade, D, Barlow, P.N, Atkinson, J.P. | | 登録日 | 2013-08-27 | | 公開日 | 2013-11-13 | | 最終更新日 | 2014-01-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Using Mutagenesis and Structural Biology to Map the Binding Site for the Plasmodium falciparum Merozoite Protein PfRh4 on the Human Immune Adherence Receptor.
J.Biol.Chem., 289, 2014
|
|
2MA8
 
 | | Solution NMR Structure of Salmonella typhimurium LT2 Secreted Protein SrfN: Northeast Structural Genomics Consortium Target StR109 | | 分子名称: | Putative secreted protein | | 著者 | Cort, J.R, Eletsky, A, Adkins, J.N, Burnet, M.C, Parish, D, Liu, K, Sukumaran, D.K, Jiang, M, Cunningham, K, Ma, T, Xiao, R, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2013-06-29 | | 公開日 | 2013-09-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and Functional Characterization of DUF1471 Domains of Salmonella Proteins SrfN, YdgH/SssB, and YahO.
Plos One, 9, 2014
|
|
6S8W
 
 | | Aromatic aminotransferase AroH (Aro8) form Aspergillus fumigatus in complex with PLP (internal aldimine) | | 分子名称: | Aromatic aminotransferase Aro8, putative, FORMIC ACID, ... | | 著者 | Giardina, G, Mirco, D, Spizzichino, S, Zelante, T, Cutruzzola, F, Romani, L, Cellini, B. | | 登録日 | 2019-07-10 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of Aspergillus fumigatus AroH, an aromatic amino acid aminotransferase
Proteins, 2021
|
|
6SHT
 
 | | Molecular structure of mouse apoferritin resolved at 2.7 Angstroms with the Glacios cryo-microscope | | 分子名称: | FE (III) ION, Ferritin heavy chain, MAGNESIUM ION | | 著者 | Hamdi, F, Tueting, C, Semchonok, D, Kyrilis, F, Meister, A, Skalidis, I, Schmidt, L, Parthier, C, Stubbs, M.T, Kastritis, P.L. | | 登録日 | 2019-08-08 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | 2.7 angstrom cryo-EM structure of vitrified M. musculus H-chain apoferritin from a compact 200 keV cryo-microscope.
Plos One, 15, 2020
|
|
3IQY
 
 | | Active site mutants of B. subtilis SecA | | 分子名称: | Protein translocase subunit secA, SULFATE ION | | 著者 | Kim, D, Hunt, J.F. | | 登録日 | 2009-08-21 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | ATPase Active-Site Electrostatic Interactions Control the Global Conformation of the 100 kDa SecA Translocase.
J.Am.Chem.Soc., 135, 2013
|
|
2M8U
 
 | | Solution structure of the Dictyostelium discodieum Myosin Light Chain, MlcC | | 分子名称: | Myosin Light Chain, MlcC | | 著者 | Liburd, J.D, Miller, E, Langelaan, D, Chitayat, S, Crawley, S.W, Cote, G.P, Smith, S.P. | | 登録日 | 2013-05-28 | | 公開日 | 2014-12-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the Single-lobe Myosin Light Chain C in Complex with the Light Chain-binding Domains of Myosin-1C Provides Insights into Divergent IQ Motif Recognition.
J.Biol.Chem., 291, 2016
|
|
2M5T
 
 | | Solution structure of the 2A proteinase from a common cold agent, human rhinovirus RV-C02, strain W12 | | 分子名称: | ZINC ION, human rhinovirus 2A proteinase | | 著者 | Lee, W, Frederick, R, Tonelli, M, Troupis, A.T, Reinin, N, Suchy, F.P, Moyer, K, Watters, K, Aceti, D, Palmenberg, A.C, Markley, J.L. | | 登録日 | 2013-03-07 | | 公開日 | 2014-03-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of the 2A Protease from a Common Cold Agent, Human Rhinovirus C2, Strain W12.
Plos One, 9, 2014
|
|
6SGF
 
 | | Molecular insight into a new low affinity xylan binding module CBM86, from the xylanolytic gut symbiont Roseburia intestinalis. | | 分子名称: | Beta-xylanase, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Ejby, M, Abou Hachem, M, Leth, M.L, Guskov, A, Slotboom, D. | | 登録日 | 2019-08-04 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.756 Å) | | 主引用文献 | Molecular insight into a new low-affinity xylan binding module from the xylanolytic gut symbiont Roseburia intestinalis.
Febs J., 287, 2020
|
|
3IFW
 
 | |
6SDU
 
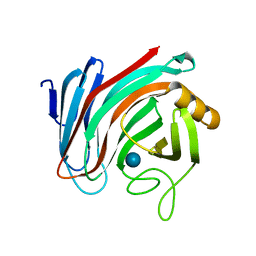 | | Xyloglucanase domain of NopAA, a type three effector from Sinorhizobium fredii in complex with cellobiose | | 分子名称: | Type III effector NopAA, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Dorival, D, Philys, S, Guintini, E, Brailly, R, de Ruyck, J, Czjzek, M, Biondi, E. | | 登録日 | 2019-07-29 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural and enzymatic characterisation of the Type III effector NopAA (=GunA) from Sinorhizobium fredii USDA257 reveals a Xyloglucan hydrolase activity.
Sci Rep, 10, 2020
|
|
2LUZ
 
 | | Solution NMR Structure of CalU16 from Micromonospora echinospora, Northeast Structural Genomics Consortium (NESG) Target MiR12 | | 分子名称: | CalU16 | | 著者 | Ramelot, T.A, Yang, Y, Lee, H, Pederson, K, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T.B, Everett, J.K, Wrobel, R.L, Bingman, C.A, Singh, S, Thorson, J.S, Prestegard, J.H, Montelione, G.T, Phillips Jr, G.N, Kennedy, M.A, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2012-06-22 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-Guided Functional Characterization of Enediyne Self-Sacrifice Resistance Proteins, CalU16 and CalU19.
Acs Chem.Biol., 9, 2014
|
|
3WI5
 
 | | Crystal structure of the Loop 7 mutant PorB from Neisseria meningitidis serogroup B | | 分子名称: | CITRATE ANION, Major outer membrane protein P.IB | | 著者 | Kattner, C, Toussi, D, Wetzler, L.M, Ruppel, N, Massari, P, Tanabe, M. | | 登録日 | 2013-09-05 | | 公開日 | 2014-01-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystallographic analysis of Neisseria meningitidis PorB extracellular loops potentially implicated in TLR2 recognition.
J.Struct.Biol., 185, 2014
|
|
7ZNR
 
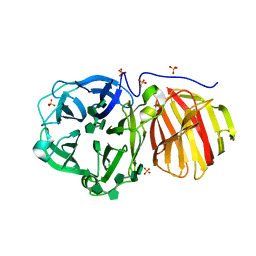 | |
7ZNS
 
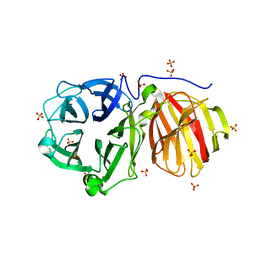 | |
2MBC
 
 | | Solution Structure of human holo-PRL-3 in complex with vanadate | | 分子名称: | Protein tyrosine phosphatase type IVA 3 | | 著者 | Jeong, K, Kang, D, Kim, J, Shin, S, Jin, B, Lee, C, Kim, E, Jeon, Y.H, Kim, Y. | | 登録日 | 2013-07-29 | | 公開日 | 2013-10-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3.
Biochemistry, 53, 2014
|
|
2MCZ
 
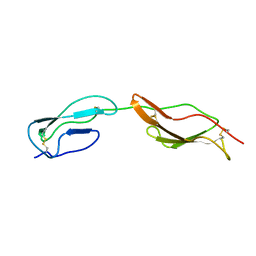 | | CR1 Sushi domains 1 and 2 | | 分子名称: | Complement receptor type 1 | | 著者 | Park, H.J, Guariento, M.J, Maciejewski, M, Hauart, R, Tham, W, Cowman, A.F, Schmidt, C.Q, Martens, H, Liszewski, K.M, Hourcade, D, Barlow, P.N, Atkinson, J.P. | | 登録日 | 2013-08-27 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Using Mutagenesis and Structural Biology to Map the Binding Site for the Plasmodium falciparum Merozoite Protein PfRh4 on the Human Immune Adherence Receptor.
J.Biol.Chem., 289, 2014
|
|
2M8G
 
 | |
