3F1X
 
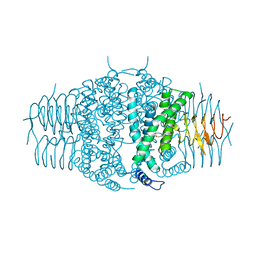 | | Three dimensional structure of the serine acetyltransferase from Bacteroides vulgatus, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET BVR62. | | 分子名称: | Serine acetyltransferase | | 著者 | Kuzin, A.P, Chen, Y, Seetharaman, J, Sahdev, S, Wang, D, Mao, L, Cunningham, K, Maglaqui, M, Xiao, R, Liu, J, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2008-10-28 | | 公開日 | 2008-11-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Three dimensional structure of the serine acetyltransferase from Bacteroides vulgatus, NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET BVR62.
To be Published
|
|
5W46
 
 | |
2KXF
 
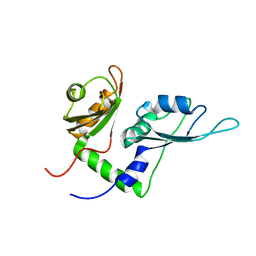 | | Solution structure of the first two RRM domains of FBP-interacting repressor (FIR) | | 分子名称: | Poly(U)-binding-splicing factor PUF60 | | 著者 | Cukier, C.D, Ramos, A, Hollingworth, D, Diaz-Moreno, I, Kelly, G. | | 登録日 | 2010-05-04 | | 公開日 | 2010-08-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular basis of FIR-mediated c-myc transcriptional control.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2KXI
 
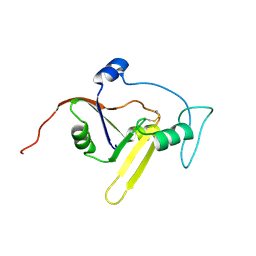 | | Solution NMR structure of the apoform of NarE (NMB1343) | | 分子名称: | Uncharacterized protein | | 著者 | Koehler, C, Carlier, L, Veggi, D, Soriani, M, Pizza, M, Boelens, R, Bonvin, A.M.J.J. | | 登録日 | 2010-05-06 | | 公開日 | 2011-03-02 | | 最終更新日 | 2018-08-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of the apoform of NarE (NMB1343)
To be Published
|
|
3F3W
 
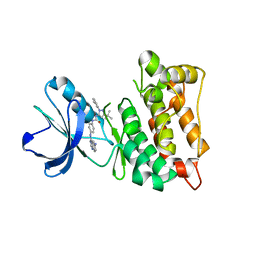 | | Drug resistant cSrc kinase domain in complex with inhibitor RL45 (Type II) | | 分子名称: | 1-{4-[(6-aminoquinazolin-4-yl)amino]phenyl}-3-[3-tert-butyl-1-(3-methylphenyl)-1H-pyrazol-5-yl]urea, Proto-oncogene tyrosine-protein kinase Src | | 著者 | Grutter, C, Kluter, S, Getlik, M, Rauh, D. | | 登録日 | 2008-10-31 | | 公開日 | 2009-06-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Hybrid compound design to overcome the gatekeeper T338M mutation in cSrc
J.Med.Chem., 52, 2009
|
|
3U8M
 
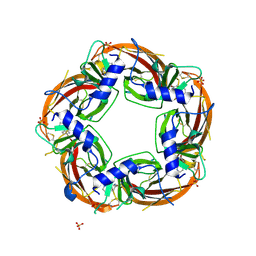 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3920 (1-(6-bromopyridin-3-yl)-1,4-diazepane) | | 分子名称: | 1-(6-bromopyridin-3-yl)-1,4-diazepane, Acetylcholine-binding protein, SULFATE ION | | 著者 | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | 登録日 | 2011-10-17 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
5W5T
 
 | | Agrobacterium tumefaciens ADP-Glucose Pyrophosphorylase bound to activator ethyl pyruvate | | 分子名称: | GLYCEROL, Glucose-1-phosphate adenylyltransferase, SULFATE ION, ... | | 著者 | Mascarenhas, R.N, Hill, B.L, Ballicora, M.A, Liu, D. | | 登録日 | 2017-06-15 | | 公開日 | 2018-11-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
2KUW
 
 | |
2KWV
 
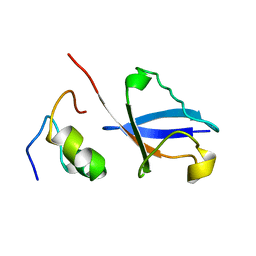 | | Solution Structure of UBM1 of murine Polymerase iota in Complex with Ubiquitin | | 分子名称: | DNA polymerase iota, Ubiquitin | | 著者 | Burschowsky, D, Rudolf, F, Rabut, G, Herrmann, T, Peter, M, Wider, G. | | 登録日 | 2010-04-20 | | 公開日 | 2010-10-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
2KWU
 
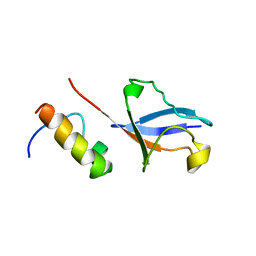 | | Solution Structure of UBM2 of murine Polymerase iota in Complex with Ubiquitin | | 分子名称: | DNA polymerase iota, Ubiquitin | | 著者 | Burschowsky, D, Rudolf, F, Rabut, G, Herrmann, T, Peter, M, Wider, G. | | 登録日 | 2010-04-19 | | 公開日 | 2010-10-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural analysis of the conserved ubiquitin-binding motifs (UBMs) of the translesion polymerase iota in complex with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
3GTK
 
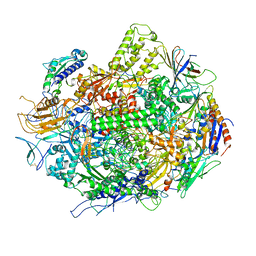 | | Backtracked RNA polymerase II complex with 18mer RNA | | 分子名称: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | 登録日 | 2009-03-27 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
2KY4
 
 | | Solution NMR structure of the PBS linker domain of phycobilisome linker polypeptide from Anabaena sp. Northeast Structural Genomics Consortium Target NsR123E | | 分子名称: | Phycobilisome linker polypeptide | | 著者 | He, Y, Eletsky, A, Mills, J.L, Lee, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Lee, H, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-05-14 | | 公開日 | 2010-07-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR structure of the PBS linker domain of phycobilisome linker polypeptide from Anabaena sp. Northeast Structural Genomics Consortium Target NsR123E
To be Published
|
|
3WAJ
 
 | |
2KE4
 
 | |
3H0M
 
 | | Structure of trna-dependent amidotransferase gatcab from aquifex aeolicus | | 分子名称: | Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, GLUTAMINE, Glutamyl-tRNA(Gln) amidotransferase subunit A, ... | | 著者 | Wu, J, Bu, W, Sheppard, K, Kitabatake, M, Soll, D, Smith, J.L. | | 登録日 | 2009-04-09 | | 公開日 | 2009-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Insights into tRNA-Dependent Amidotransferase Evolution and Catalysis from the Structure of the Aquifex aeolicus Enzyme
J.Mol.Biol., 391, 2009
|
|
3WEV
 
 | | Crystal structure of the Schiff base intermediate of L-Lys epsilon-oxidase from Marinomonas mediterranea with L-Lys | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, L-lysine 6-oxidase, ... | | 著者 | Okazaki, S, Nakano, S, Matsui, D, Akaji, S, Inagaki, K, Asano, Y. | | 登録日 | 2013-07-12 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | X-Ray crystallographic evidence for the presence of the cysteine tryptophylquinone cofactor in L-lysine {varepsilon}-oxidase from Marinomonas mediterranea
J.Biochem., 154, 2013
|
|
2KVS
 
 | | NMR Solution Structure of Q7A1E8 protein from Staphylococcus aureus: Northeast Structural Genomics Consortium target: ZR215 | | 分子名称: | Uncharacterized protein MW0776 | | 著者 | Swapna, G.V.T, Montelione, A.F, Wang, D, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Rost, B, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-03-27 | | 公開日 | 2010-04-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Solution Structure of Q7A1E8 protein from Staphylococcus aureus: Northeast Structural Genomics Consortium target: ZR215
To be Published
|
|
2HR9
 
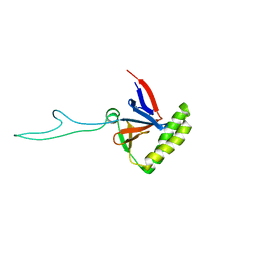 | |
3WDB
 
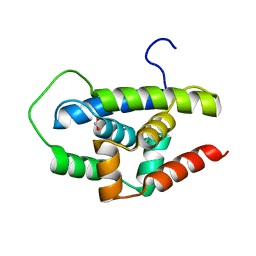 | |
3GRO
 
 | | Human palmitoyl-protein thioesterase 1 | | 分子名称: | Palmitoyl-protein thioesterase 1, UNKNOWN ATOM OR ION | | 著者 | Dobrovetsky, E, Seitova, A, Tong, Y, Tempel, W, Dong, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Cossar, D, Park, H, Structural Genomics Consortium (SGC) | | 登録日 | 2009-03-26 | | 公開日 | 2009-04-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Human palmitoyl-protein thioesterase 1
To be Published
|
|
3WH2
 
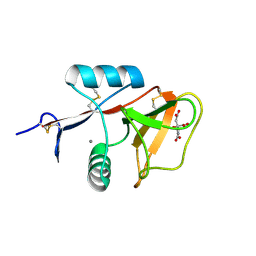 | | Human Mincle in complex with citrate | | 分子名称: | C-type lectin domain family 4 member E, CALCIUM ION, CITRATE ANION | | 著者 | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | 登録日 | 2013-08-21 | | 公開日 | 2013-10-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3WHD
 
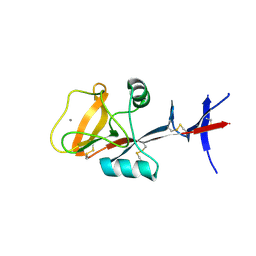 | | C-type lectin, human MCL | | 分子名称: | C-type lectin domain family 4 member D, CALCIUM ION | | 著者 | Furukawa, A, Kamishikiryo, J, Mori, D, Toyonaga, K, Okabe, Y, Toji, A, Kanda, R, Miyake, Y, Ose, T, Yamasaki, S, Maenaka, K. | | 登録日 | 2013-08-24 | | 公開日 | 2013-10-23 | | 最終更新日 | 2013-12-11 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural analysis for glycolipid recognition by the C-type lectins Mincle and MCL
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3GSR
 
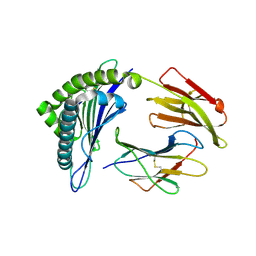 | | Crystal structure of the binary complex between HLA-A2 and HCMV NLV-M5V peptide variant | | 分子名称: | Beta-2-microglobulin, HCMV pp65 fragment 495-503, variant M5V (NLVPVVATV), ... | | 著者 | Reiser, J.-B, Saulquin, X, Gras, S, Debeaupuis, E, Echasserieau, K, Kissenpfennig, A, Legoux, F, Chouquet, A, Le Gorrec, M, Machillot, P, Neveu, B, Thielens, N, Malissen, B, Bonneville, M, Housset, D. | | 登録日 | 2009-03-27 | | 公開日 | 2009-08-04 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope.
J.Immunol., 183, 2009
|
|
3H6Q
 
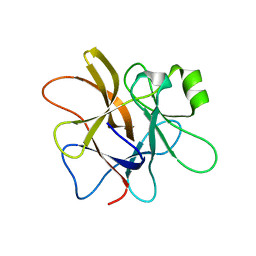 | | Macrocypin, a beta-trefoil cysteine protease inhibitor | | 分子名称: | Macrocypin 1a | | 著者 | Renko, M, Sabotic, J, Brzin, J, Turk, D. | | 登録日 | 2009-04-23 | | 公開日 | 2009-10-20 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.643 Å) | | 主引用文献 | Versatile loops in mycocypins inhibit three protease families.
J.Biol.Chem., 285, 2010
|
|
3GTP
 
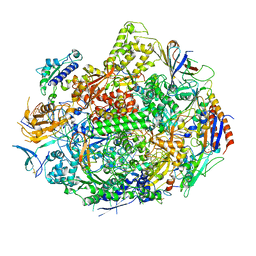 | | Backtracked RNA polymerase II complex with 24mer RNA | | 分子名称: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | 著者 | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | 登録日 | 2009-03-27 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
