6BES
 
 | | Solution structure of de novo macrocycle design11_ss | | 分子名称: | (DAL)Q(DPR)(DCY)(DLY)DS(DTY)(DCY)P(DSN) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-10-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6B3O
 
 | | Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion | | 分子名称: | Spike glycoprotein | | 著者 | Walls, A.C, Tortorici, M.A, Snijder, J, Xiong, X, Bosch, B.J, Rey, F.A, Veesler, D. | | 登録日 | 2017-09-22 | | 公開日 | 2017-10-04 | | 最終更新日 | 2020-01-01 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1LFW
 
 | | Crystal structure of pepV | | 分子名称: | 3-[(1-AMINO-2-CARBOXY-ETHYL)-HYDROXY-PHOSPHINOYL]-2-METHYL-PROPIONIC ACID, ZINC ION, pepV | | 著者 | Jozic, D, Bourenkow, G, Bartunik, H, Scholze, H, Dive, V, Henrich, B, Huber, R, Bode, W, Maskos, K. | | 登録日 | 2002-04-12 | | 公開日 | 2002-10-23 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of the Dinuclear Zinc Aminopeptidase PepV from Lactobacillus delbrueckii Unravels Its Preference for Dipeptides
Structure, 10, 2002
|
|
6ZFM
 
 | | Structure of alpha-Cobratoxin with a peptide inhibitor | | 分子名称: | 3-[2-[2-[2-[2-[2-(2-azanylethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]propan-1-ol, Alpha-cobratoxin, PENTAETHYLENE GLYCOL, ... | | 著者 | Kiontke, S, Kummel, D. | | 登録日 | 2020-06-17 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Peptide Inhibitors of the alpha-Cobratoxin-Nicotinic Acetylcholine Receptor Interaction.
J.Med.Chem., 63, 2020
|
|
6IBS
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with boronic inhibitor cpd 6 | | 分子名称: | CALCIUM ION, Metallo-beta-lactamase type 2, ZINC ION, ... | | 著者 | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | 登録日 | 2018-11-30 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6BEF
 
 | | Crystal structure of VACV D13 in complex with 3-formyl rifamycin SV | | 分子名称: | (2S,12Z,14E,16S,17S,18R,19R,20R,21S,22R,23S,24E)-8-formyl-5,6,9,17,19-pentahydroxy-23-methoxy-2,4,12,16,18,20,22-heptam ethyl-1,11-dioxo-1,2-dihydro-2,7-(epoxypentadeca[1,11,13]trienoimino)naphtho[2,1-b]furan-21-yl acetate, 1,2-ETHANEDIOL, FORMIC ACID, ... | | 著者 | Garriga, D, Accurso, C, Coulibaly, F. | | 登録日 | 2017-10-25 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Structural basis for the inhibition of poxvirus assembly by the antibiotic rifampicin.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6ZIR
 
 | | CRYSTAL STRUCTURE OF NRAS (C118S) IN COMPLEX WITH GDP AND COMPOUND 18 | | 分子名称: | (3~{S})-3-[2-[(dimethylamino)methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase NRas, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Kessler, D, Fischer, G, Boettcher, J. | | 登録日 | 2020-06-26 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Drugging all RAS isoforms with one pocket.
Future Med Chem, 12, 2020
|
|
1L0A
 
 | | DOWNSTREAM REGULATOR TANK BINDS TO THE CD40 RECOGNITION SITE ON TRAF3 | | 分子名称: | TNF receptor associated factor 3, TRAF family member-associated NF-kappa-b activator | | 著者 | Li, C, Ni, C.-Z, Havert, M.L, Cabezas, E, He, J, Kaiser, D, Reed, J.C, Satterthwait, A.C, Cheng, G, Ely, K.R. | | 登録日 | 2002-02-08 | | 公開日 | 2002-04-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Downstream regulator TANK binds to the CD40 recognition site on TRAF3.
Structure, 10, 2002
|
|
6ZIW
 
 | | The IRAK3 Pseudokinase Domain Bound To ATPgammaS | | 分子名称: | Interleukin-1 receptor-associated kinase 3, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SULFATE ION | | 著者 | Mathea, S, Chatterjee, D, Preuss, F, Kraemer, A, Knapp, S. | | 登録日 | 2020-06-26 | | 公開日 | 2020-07-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | The IRAK3 Pseudokinase Domain Bound To ATPgammaS
To Be Published
|
|
6ZJ0
 
 | | CRYSTAL STRUCTURE OF HRAS-G12D IN COMPLEX WITH GCP AND COMPOUND 18 | | 分子名称: | (3~{S})-3-[2-[(dimethylamino)methyl]-1~{H}-indol-3-yl]-5-oxidanyl-2,3-dihydroisoindol-1-one, GTPase HRas, MAGNESIUM ION, ... | | 著者 | Kessler, D, Fischer, G, Boettcher, J. | | 登録日 | 2020-06-26 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.763 Å) | | 主引用文献 | Drugging all RAS isoforms with one pocket.
Future Med Chem, 12, 2020
|
|
6I1N
 
 | | Plasmodium falciparum spermidine synthase in complex with N-(3-aminopropyl)-trans-cyclohexane-1,4-diamine | | 分子名称: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, GLYCEROL, Spermidine synthase, ... | | 著者 | Sprenger, J, Coertzen, D, Persson, L, Carey, J, Birkholtz, L.M, Louw, B.I. | | 登録日 | 2018-10-29 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.85000134 Å) | | 主引用文献 | Plasmodium falciparum spermidine synthase in complex with N-(3-aminopropyl)-trans-cyclohexane-1,4-diamine
To Be Published
|
|
4QXX
 
 | |
6ZK9
 
 | | Peripheral domain of open complex I during turnover | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | 著者 | Kampjut, D, Sazanov, L.A. | | 登録日 | 2020-06-30 | | 公開日 | 2020-10-07 | | 最終更新日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6ZKN
 
 | | Complex I inhibited by rotenone, open3 | | 分子名称: | (2R,6aS,12aS)-8,9-dimethoxy-2-(prop-1-en-2-yl)-1,2,12,12a-tetrahydrofuro[2',3':7,8][1]benzopyrano[2,3-c][1]benzopyran-6(6aH)-one, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ... | | 著者 | Kampjut, D, Sazanov, L.A. | | 登録日 | 2020-06-30 | | 公開日 | 2020-10-07 | | 最終更新日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6BF3
 
 | | Solution structure of de novo macrocycle design7.3a | | 分子名称: | QDP(DPR)K(2TL)(DAS) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BE7
 
 | | Solution structure of de novo macrocycle Design8.1 | | 分子名称: | DDPT(DPR)(DAR)Q(DGN) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-24 | | 公開日 | 2018-01-03 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6ZKQ
 
 | | Native complex I, open2 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE MONOPHOSPHATE, ... | | 著者 | Kampjut, D, Sazanov, L.A. | | 登録日 | 2020-06-30 | | 公開日 | 2020-10-07 | | 最終更新日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6YT6
 
 | |
6I1H
 
 | | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with meropenem | | 分子名称: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein,Penicillin-binding protein | | 著者 | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | 登録日 | 2018-10-28 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Crystal structure of TP domain from Chlamydia trachomatis Penicillin-Binding Protein 3 in complex with meropenem
To Be Published
|
|
6ZI8
 
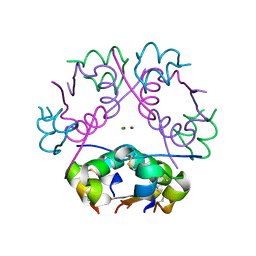 | | X-ray diffraction structure of bovine insulin at 2.3 A resolution | | 分子名称: | CHLORIDE ION, Insulin, ZINC ION | | 著者 | Housset, D, Ling, W.L, Bacia-Verloop, M, Zander, U, McCarthy, A.A, Schoehn, G. | | 登録日 | 2020-06-25 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Statistically correcting dynamical electron scattering improves the refinement of protein nanocrystals, including charge refinement of coordinated metals.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1L7Y
 
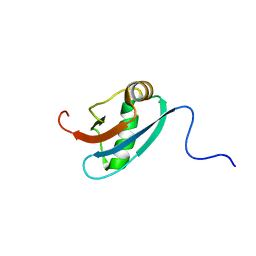 | | Solution NMR Structure of C. elegans Protein ZK652.3. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET WR41. | | 分子名称: | HYPOTHETICAL PROTEIN ZK652.3 | | 著者 | Cort, J.R, Chiang, Y, Zheng, D, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2002-03-18 | | 公開日 | 2002-08-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of conserved eukaryotic protein ZK652.3 from C. elegans: a ubiquitin-like fold.
Proteins, 48, 2002
|
|
6ZKC
 
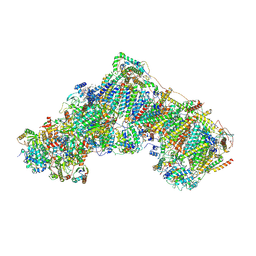 | | Complex I during turnover, closed | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | 著者 | Kampjut, D, Sazanov, L.A. | | 登録日 | 2020-06-30 | | 公開日 | 2020-10-07 | | 最終更新日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6ZKR
 
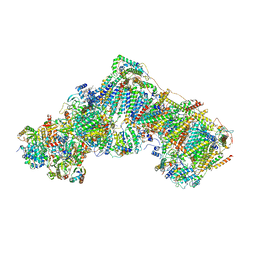 | | Native complex I, open3 | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE MONOPHOSPHATE, ... | | 著者 | Kampjut, D, Sazanov, L.A. | | 登録日 | 2020-06-30 | | 公開日 | 2020-10-07 | | 最終更新日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6YVS
 
 | | FOCAL ADHESION KINASE CATALYTIC DOMAIN IN COMPLEX WITH 5-{4-[(Pyridin-3-ylmethyl)-amino]-5-trifluoromethyl-pyrimidin-2-ylamino}-1,3-dihydro-indol-2-one | | 分子名称: | 5-[[4-(pyridin-3-ylmethylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino]-1,3-dihydroindol-2-one, Focal adhesion kinase 1, SULFATE ION | | 著者 | Musil, D, Heinrich, T, Amaral, M. | | 登録日 | 2020-04-28 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structure-kinetic relationship reveals the mechanism of selectivity of FAK inhibitors over PYK2.
Cell Chem Biol, 28, 2021
|
|
6I53
 
 | | Cryo-EM structure of the human synaptic alpha1-beta3-gamma2 GABAA receptor in complex with Megabody38 in a lipid nanodisc | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | 著者 | Laverty, D, Desai, R, Uchanski, T, Masiulis, S, Wojciech, J.S, Malinauskas, T, Zivanov, J, Pardon, E, Steyaert, J, Miller, K.W, Aricescu, A.R. | | 登録日 | 2018-11-12 | | 公開日 | 2019-01-02 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of the human alpha 1 beta 3 gamma 2 GABAAreceptor in a lipid bilayer.
Nature, 565, 2019
|
|
