2RLP
 
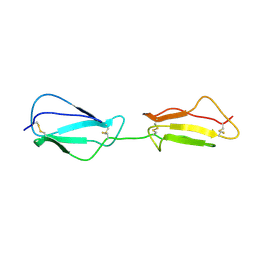 | | NMR structure of CCP modules 1-2 of complement factor H | | 分子名称: | Complement factor H | | 著者 | Hocking, H.G, Herbert, A.P, Pangburn, M.K, Kavanagh, D, Barlow, P.N, Uhrin, D. | | 登録日 | 2007-07-28 | | 公開日 | 2008-02-19 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the N-terminal region of complement factor H and conformational implications of disease-linked sequence variations.
J.Biol.Chem., 283, 2008
|
|
1EVV
 
 | |
1BFD
 
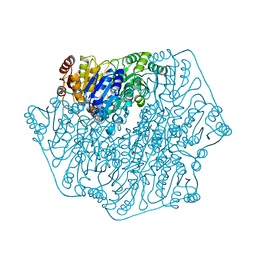 | | BENZOYLFORMATE DECARBOXYLASE FROM PSEUDOMONAS PUTIDA | | 分子名称: | BENZOYLFORMATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | 著者 | Hasson, M.S, Muscate, A, Mcleish, M.J, Polovnikova, L.S, Gerlt, J.A, Kenyon, G.L, Petsko, G.A, Ringe, D. | | 登録日 | 1998-04-30 | | 公開日 | 1998-06-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The crystal structure of benzoylformate decarboxylase at 1.6 A resolution: diversity of catalytic residues in thiamin diphosphate-dependent enzymes.
Biochemistry, 37, 1998
|
|
1EEH
 
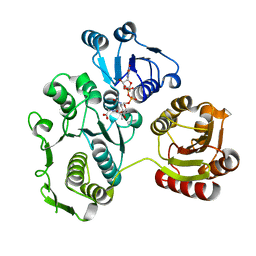 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | 分子名称: | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE | | 著者 | Bertrand, J.A, Fanchon, E, Martin, L, Chantalat, L, Auger, G, Blanot, D, van Heijenoort, J, Dideberg, O. | | 登録日 | 2000-01-31 | | 公開日 | 2001-01-17 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | "Open" structures of MurD: domain movements and structural similarities with folylpolyglutamate synthetase.
J.Mol.Biol., 301, 2000
|
|
1S52
 
 | | Thr24Val Bacteriorhodopsin | | 分子名称: | RETINAL, bacteriorhodopsin | | 著者 | Yohannan, S, Faham, S, Yang, D, Grosfeld, D, Chamberlain, A.K, Bowie, J.U. | | 登録日 | 2004-01-19 | | 公開日 | 2004-03-02 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A C(alpha)-H.O Hydrogen Bond in a Membrane Protein Is Not Stabilizing
J.Am.Chem.Soc., 126, 2004
|
|
7ND4
 
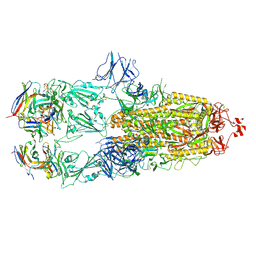 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-88 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-88 Fab heavy chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND3
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-40 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-40 heavy chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND8
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-384 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-384 Fab heavy chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7NDC
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein (all RBD down) in complex with COVOX-159 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-159 Fab light chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND7
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-316 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND9
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein (one RBD up) in complex with COVOX-253H55L Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7NDD
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein (one RBD up) in complex with COVOX-159 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-159 Fab heavy chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7NDA
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein (all RBD down) in complex with COVOX-253H55L Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND5
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-150 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-150 Fab heavy chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND6
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-40 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 Fab heavy chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (7.3 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7NDB
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-253H165L Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
2RKX
 
 | | The 3D structure of chain D, cyclase subunit of imidazoleglycerol_evolvedcerolphosphate synthase | | 分子名称: | Cyclase subunit of imidazoleglycerol_evolvedcerolphosphate synthase | | 著者 | Tawfik, D, Khersonsky, O, Albeck, S, Dym, O, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2007-10-18 | | 公開日 | 2008-03-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Kemp elimination catalysts by computational enzyme design.
Nature, 453, 2008
|
|
1R4T
 
 | | Solution structure of exoenzyme S | | 分子名称: | exoenzyme S | | 著者 | Langdon, G.M, Leitner, D, Labudde, D, Kuhne, R, Schmieder, P, Aktories, K, Oschkinat, H.O, Schmidt, G. | | 登録日 | 2003-10-08 | | 公開日 | 2005-04-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the N-terminal GTPase activating domain of Pseudomonas aeruginosa exoenzyme S
To be Published
|
|
1BD0
 
 | |
1JFW
 
 | |
7NDV
 
 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001888. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[4-(trifluoromethyl)phenoxy]piperidine, Acetylcholine-binding protein, ... | | 著者 | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | 登録日 | 2021-02-02 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Discovery of fragments inducing conformational effects in dynamic proteins using a second-harmonic generation biosensor
RSC Advances, 11, 2021
|
|
7W3F
 
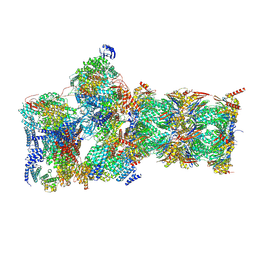 | | Structure of USP14-bound human 26S proteasome in substrate-engaged state ED1_USP14 | | 分子名称: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | 著者 | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | 登録日 | 2021-11-25 | | 公開日 | 2022-05-04 | | 最終更新日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
7W38
 
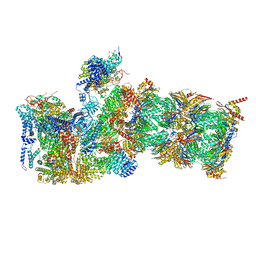 | | Structure of USP14-bound human 26S proteasome in state EA2.0_UBL | | 分子名称: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | 著者 | Zhang, S, Zou, S, Yin, D, Wu, Z, Mao, Y. | | 登録日 | 2021-11-25 | | 公開日 | 2022-05-04 | | 最終更新日 | 2022-06-01 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | USP14-regulated allostery of the human proteasome by time-resolved cryo-EM.
Nature, 605, 2022
|
|
6O4J
 
 | | Amyloid Beta KLVFFAENVGS 16-26 D23N Iowa mutation | | 分子名称: | Amyloid-beta precursor protein | | 著者 | Griner, S.L, Sawaya, M.R, Rodriguez, J.A, Cascio, D, Gonen, T. | | 登録日 | 2019-02-28 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (1.402 Å) | | 主引用文献 | Structure based inhibitors of Amyloid Beta core suggest a common interface with Tau.
Elife, 8, 2019
|
|
5LJM
 
 | | Structure of SPATA2 PUB domain | | 分子名称: | GLYCEROL, Spermatogenesis-associated protein 2 | | 著者 | Elliott, P.R, Komander, D. | | 登録日 | 2016-07-18 | | 公開日 | 2016-08-24 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.454 Å) | | 主引用文献 | SPATA2 Links CYLD to LUBAC, Activates CYLD, and Controls LUBAC Signaling.
Mol.Cell, 63, 2016
|
|
