2QW0
 
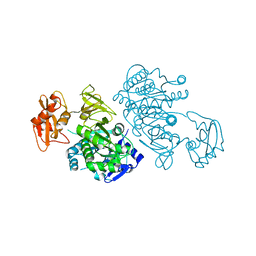 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, I303A mutation, bound to 3,4 Dichlorobenzoate | | Descriptor: | 3,4-dichlorobenzoate, 4-Chlorobenzoate CoA Ligase | | Authors: | Wu, R, Reger, A.S, Cao, J, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2007-08-09 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Rational redesign of the 4-chlorobenzoate binding site of 4-chlorobenzoate: coenzyme a ligase for expanded substrate range.
Biochemistry, 46, 2007
|
|
7ZUF
 
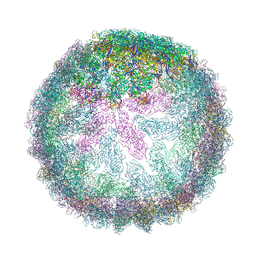 | | Saccharomyces cerevisiae L-BC virus, open particle, C5 reconstruction | | Descriptor: | Major capsid protein | | Authors: | Grybchuk, D, Prochazkova, M, Fuzik, T, Konovalovas, A, Serva, S, Yurchenko, V, Plevka, P. | | Deposit date: | 2022-05-12 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structures of L-BC virus and its open particle provide insight into Totivirus capsid assembly.
Commun Biol, 5, 2022
|
|
2Q9X
 
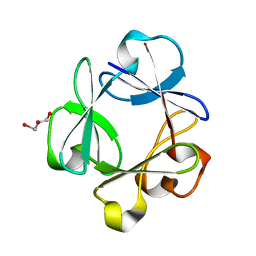 | | Crystal structure of highly stable mutant Q40P/S47I/H93G of human fibroblast growth factor-1 | | Descriptor: | GLYCEROL, Heparin-binding growth factor 1 | | Authors: | Szlachcic, A, Zakrzewska, M, Krowarsch, D, Os, V, Helland, R, Otlewski, J. | | Deposit date: | 2007-06-14 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of highly stable mutant Q40P/S47I/H93G of human fibroblast growth factor-1
To be Published
|
|
7ZMD
 
 | |
7ZMC
 
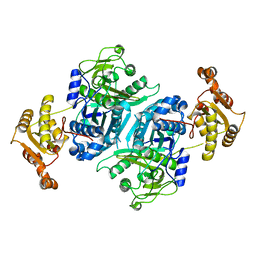 | |
7ZMA
 
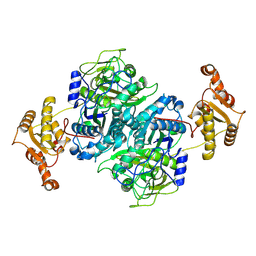 | |
2AHQ
 
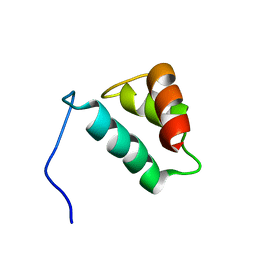 | |
2F3W
 
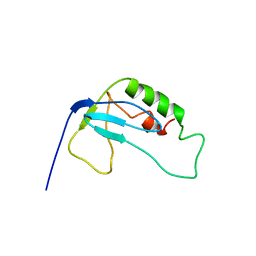 | | solution structure of 1-110 fragment of staphylococcal nuclease in 2M TMAO | | Descriptor: | Thermonuclease | | Authors: | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | Deposit date: | 2005-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
1N0X
 
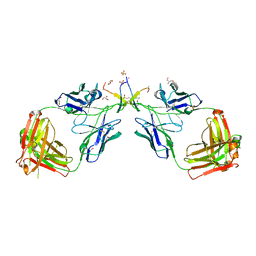 | | Crystal Structure of a Broadly Neutralizing Anti-HIV-1 Antibody in Complex with a Peptide Mimotope | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, B2.1 peptide, GLYCEROL, ... | | Authors: | Saphire, E.O, Montero, M, Menendez, A, Irving, M.B, Zwick, M.B, Parren, P.W.H.I, Burton, D.R, Scott, J.K, Wilson, I.A. | | Deposit date: | 2002-10-15 | | Release date: | 2004-04-13 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Broadly Neutralizing Anti-HIV-1 Antibody in Complex with a Peptide Mimotope
To be Published
|
|
4QMH
 
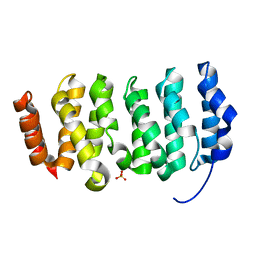 | | The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array | | Descriptor: | LP04448p, SULFATE ION | | Authors: | Fox, J.C, Howard, A.E, Currie, J.D, Rogers, S.L, Slep, K.C. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array.
Mol.Biol.Cell, 25, 2014
|
|
2QTL
 
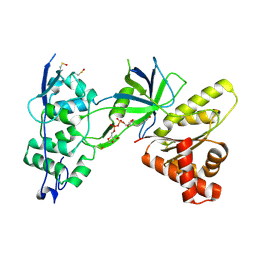 | | Crystal Structure of the FAD-containing FNR-like Module of Human Methionine Synthase Reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methionine synthase reductase | | Authors: | Wolthers, K.R, Lou, X, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2007-08-02 | | Release date: | 2007-11-13 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Coenzyme Binding to Human Methionine Synthase Reductase Revealed through the Crystal Structure of the FNR-like Module and Isothermal Titration Calorimetry
Biochemistry, 46, 2007
|
|
1XOB
 
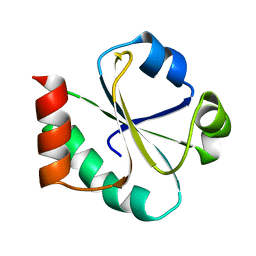 | | THIOREDOXIN (REDUCED DITHIO FORM), NMR, 20 STRUCTURES | | Descriptor: | THIOREDOXIN | | Authors: | Jeng, M.-F, Campbell, A.P, Begley, T, Holmgren, A, Case, D.A, Wright, P.E, Dyson, H.J. | | Deposit date: | 1995-11-28 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin.
Structure, 2, 1994
|
|
2QWT
 
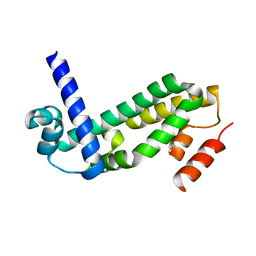 | | Crystal structure of the TetR transcription regulatory protein from Mycobacterium vanbaalenii | | Descriptor: | Transcriptional regulator, TetR family | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Adams, J, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-10 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the TetR transcription regulatory protein from Mycobacterium vanbaalenii.
To be Published
|
|
2QU8
 
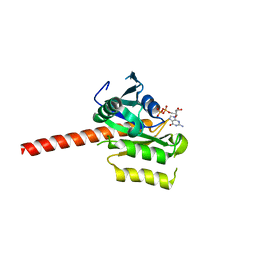 | | Crystal structure of putative nucleolar GTP-binding protein 1 PFF0625w from Plasmodium falciparum | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Putative nucleolar GTP-binding protein 1 | | Authors: | Wernimont, A.K, Lew, J, Lin, Y.H, Kozieradzki, I, Zhao, Y, Ravichandran, M, Shapiro, M, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Sukumar, D, Hassanali, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-08-03 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of putative nucleolar GTP-binding protein 1 PFF0625w from Plasmodium falciparum.
To be Published
|
|
2Y2Z
 
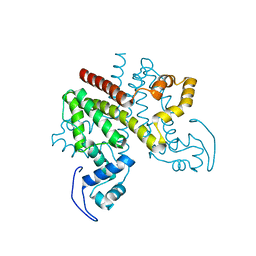 | | ligand-free form of TetR-like repressor SimR | | Descriptor: | PUTATIVE REPRESSOR SIMREG2 | | Authors: | Le, T.B.K, Stevenson, C.E.M, Fiedler, H.-P, Maxwell, A, Lawson, D.M, Buttner, M.J. | | Deposit date: | 2010-12-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Tetr-Like Simocyclinone Efflux Pump Repressor, Simr, and the Mechanism of Ligand-Mediated Derepression.
J.Mol.Biol., 408, 2011
|
|
2XVR
 
 | | Phage T7 empty mature head shell | | Descriptor: | MAJOR CAPSID PROTEIN 10A | | Authors: | Ionel, A, Velazquez-Muriel, J.A, Luque, D, Cuervo, A, Caston, J.R, Valpuesta, J.M, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2010-10-28 | | Release date: | 2010-12-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Molecular Rearrangements Involved in the Capsid Shell Maturation of Bacteriophage T7.
J.Biol.Chem., 286, 2011
|
|
2NBH
 
 | |
2W8V
 
 | | SPT with PLP, N100W | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE PALMITOYLTRANSFERASE | | Authors: | Raman, M.C.C, Johnson, K.A, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2009-01-19 | | Release date: | 2009-01-27 | | Last modified: | 2015-11-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The External-Aldimine Form of Serine Palmitoyltranserase; Structural, Kinetic and Spectroscopic Analysis of the Wild-Type Enzyme and Hsan1 Mutant Mimics.
J.Biol.Chem., 284, 2009
|
|
2OI3
 
 | | NMR Structure Analysis of the Hematopoetic Cell Kinase SH3 Domain complexed with an artificial high affinity ligand (PD1) | | Descriptor: | Tyrosine-protein kinase HCK, artificial peptide PD1 | | Authors: | Schmidt, H, Stoldt, M, Hoffmann, S, Tran, T, Willbold, D. | | Deposit date: | 2007-01-10 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Hck SH3 Domain Ligand
Complex Reveals Novel Interaction Modes
J.Mol.Biol., 365, 2007
|
|
1XMT
 
 | | X-ray structure of gene product from arabidopsis thaliana at1g77540 | | Descriptor: | BROMIDE ION, putative acetyltransferase | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
1LR1
 
 | | Solution Structure of the Oligomerization Domain of the Bacterial Chromatin-Structuring Protein H-NS | | Descriptor: | dna-binding protein h-ns | | Authors: | Esposito, D, Petrovic, A, Harris, R, Ono, S, Eccleston, J, Mbabaali, A, Haq, I, Higgins, C.F, Hinton, J.C.D, Driscoll, P.C, Ladbury, J.E. | | Deposit date: | 2002-05-14 | | Release date: | 2003-01-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | H-NS Oligomerization Domain Structure Reveals the Mechanism for High Order
Self-association of the Intact Protein
J.Mol.Biol., 324, 2002
|
|
5Q17
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-({5-bromo-2-oxo-1'-[(thiophen-2-yl)sulfonyl]spiro[indole-3,4'-piperidin]-1(2H)-yl}methyl)benzoic acid, Bile acid receptor, Nuclear receptor coactivator peptide SRC2 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q10
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N-[3-(acetylamino)phenyl]-4-chloro-N-[(1S)-1-cyclohexyl-2-(cyclohexylamino)-2-oxoethyl]benzamide | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
2VUJ
 
 | | Environmentally isolated GH11 xylanase | | Descriptor: | GH11 XYLANASE, GLYCEROL | | Authors: | Dumon, C, Varvak, A, Wall, M.A, Flint, J.E, Lewis, R.J, Lakey, J.H, Luginbuhl, P, Healey, S, Todaro, T, DeSantis, G, Sun, M, Parra-Gessert, L, Tan, X, Weiner, D.P, Gilbert, H.J. | | Deposit date: | 2008-05-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering Hyperthermostability Into a Gh11 Xylanase is Mediated by Subtle Changes to Protein Structure.
J.Biol.Chem., 283, 2008
|
|
1XBL
 
 | | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | | Descriptor: | DNAJ | | Authors: | Pellecchia, M, Szyperski, T, Wall, D, Georgopoulos, C, Wuthrich, K. | | Deposit date: | 1996-10-07 | | Release date: | 1997-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone.
J.Mol.Biol., 260, 1996
|
|
