1U42
 
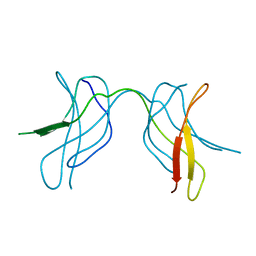 | | Crystal structure of MLAM mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
4W7A
 
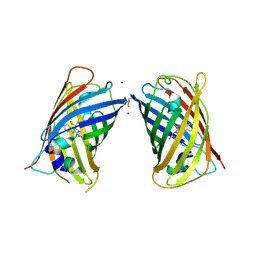 | | Crystal Structure of Full-Length Split GFP Mutant D21H/K26C Disulfide and Metal-Mediated Dimer, P 21 21 21 Space Group, Form 4 | | Descriptor: | COPPER (II) ION, fluorescent protein D21H/K26C | | Authors: | Leibly, D.J, Waldo, G.S, Yeates, T.O. | | Deposit date: | 2014-08-21 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.603 Å) | | Cite: | A Suite of Engineered GFP Molecules for Oligomeric Scaffolding.
Structure, 23, 2015
|
|
1CZR
 
 | | COMPARISONS OF WILD TYPE AND MUTANT FLAVODOXINS FROM ANACYSTIS NIDULANS. STRUCTURAL DETERMINANTS OF THE REDOX POTENTIALS. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Hoover, D.M, Drennan, C.L, Metzger, A.L, Osborne, C, Weber, C.H, Pattridge, K.A, Ludwig, M.L. | | Deposit date: | 1999-09-07 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials.
J.Mol.Biol., 294, 1999
|
|
1D04
 
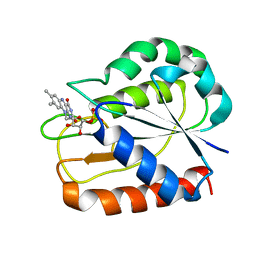 | | COMPARISONS OF WILD TYPE AND MUTANT FLAVODOXINS FROM ANACYSTIS NIDULANS. STRUCTURAL DETERMINANTS OF THE REDOX POTENTIALS. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Hoover, D.M, Drennan, C.L, Metzger, A.L, Osborne, C, Weber, C.H, Pattridge, K.A, Ludwig, M.L. | | Deposit date: | 1999-09-08 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials.
J.Mol.Biol., 294, 1999
|
|
6QYM
 
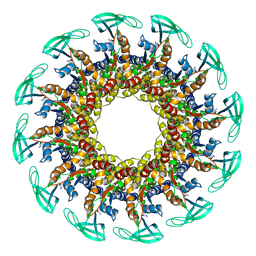 | |
8BTN
 
 | | Crystal structure of BcThsB | | Descriptor: | TIR domain-containing protein | | Authors: | Tamulaitiene, G, Sabonis, D. | | Deposit date: | 2022-11-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Activation of Thoeris antiviral system via SIR2 effector filament assembly.
Nature, 627, 2024
|
|
8BVC
 
 | |
6QSL
 
 | |
1TOE
 
 | | Unliganded structure of Hexamutant + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, SULFATE ION | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
1U6G
 
 | | Crystal Structure of The Cand1-Cul1-Roc1 Complex | | Descriptor: | Cullin homolog 1, RING-box protein 1, TIP120 protein, ... | | Authors: | Goldenberg, S.J, Shumway, S.D, Cascio, T.C, Garbutt, K.C, Liu, J, Xiong, Y, Zheng, N. | | Deposit date: | 2004-07-29 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases
Cell(Cambridge,Mass.), 119, 2004
|
|
1CJW
 
 | | SEROTONIN N-ACETYLTRANSFERASE COMPLEXED WITH A BISUBSTRATE ANALOG | | Descriptor: | COA-S-ACETYL TRYPTAMINE, PROTEIN (SEROTONIN N-ACETYLTRANSFERASE) | | Authors: | Hickman, A.B, Namboodiri, M.A.A, Klein, D.C, Dyda, F. | | Deposit date: | 1999-04-19 | | Release date: | 1999-05-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of ordered substrate binding by serotonin N-acetyltransferase: enzyme complex at 1.8 A resolution with a bisubstrate analog.
Cell(Cambridge,Mass.), 97, 1999
|
|
6QUE
 
 | |
6QYG
 
 | |
5J8D
 
 | | Structure of nitroreductase from E. cloacae complexed with nicotinic acid adenine dinucleotide | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID ADENINE DINUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2016-04-07 | | Release date: | 2017-05-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism-Informed Refinement Reveals Altered Substrate-Binding Mode for Catalytically Competent Nitroreductase.
Structure, 25, 2017
|
|
6QXV
 
 | | Pink beam serial crystallography: Proteinase K, 1 us exposure, 1585 patterns merged (2 chips) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Proteinase K, ... | | Authors: | Tolstikova, A, Oberthuer, D, Meents, A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | 1 kHz fixed-target serial crystallography using a multilayer monochromator and an integrating pixel detector.
Iucrj, 6, 2019
|
|
8C3X
 
 | |
6YUU
 
 | | Crystal structure of M. tuberculosis InhA inhibited by SKTS1 | | Descriptor: | 6-[4-(4-hexyl-2-oxidanyl-phenoxy)phenoxy]pyridin-2-ol, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Schiebel, J, Kehrein, J, Le, T.A, Davoodi, S, Merget, B, Weinrich, J.D, Tonge, P.J, Engels, B, Sotriffer, C.A, Kisker, C. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A Long Residence Time Enoyl-Reductase Inhibitor Explores an Extended Binding Region with Isoenzyme-Dependent Tautomer Adaptation and Differential Substrate-Binding Loop Closure.
Acs Infect Dis., 7, 2021
|
|
1KHE
 
 | | PEPCK complex with nonhydrolyzable GTP analog, MAD data | | Descriptor: | MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Phosphoenolpyruvate Carboxykinase, ... | | Authors: | Dunten, P, Belunis, C, Crowther, R, Hollfelder, K, Kammlott, U, Levin, W, Michel, H, Ramsey, G.B, Swain, A, Weber, D, Wertheimer, S.J. | | Deposit date: | 2001-11-29 | | Release date: | 2002-02-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site.
J.Mol.Biol., 316, 2002
|
|
6QYF
 
 | |
6QYJ
 
 | |
1KKE
 
 | |
1U5Z
 
 | | The Crystal structure of murine APRIL, pH 8.5 | | Descriptor: | NICKEL (II) ION, Tumor necrosis factor ligand superfamily member 13 | | Authors: | Wallweber, H.J, Compaan, D.M, Starovasnik, M.A, Hymowitz, S.G. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of A Proliferation-inducing Ligand, APRIL.
J.Mol.Biol., 343, 2004
|
|
6QTI
 
 | |
8COA
 
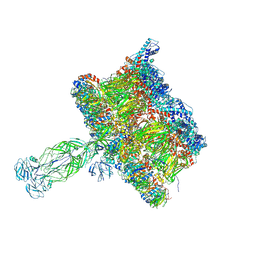 | | in situ Subtomogram average of Immature Rotavirus TLP spike | | Descriptor: | Intermediate capsid protein VP6, Outer capsid glycoprotein VP7, Outer capsid protein VP4 | | Authors: | Shah, P.N.M, Stuart, D.I. | | Deposit date: | 2023-02-27 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Characterization of the rotavirus assembly pathway in situ using cryoelectron tomography.
Cell Host Microbe, 31, 2023
|
|
6YX8
 
 | | The structure of allophycocyanin from cyanobacterium Nostoc sp. WR13, the C2221 crystal form. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Patel, H.M, Roszak, A.W, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.831 Å) | | Cite: | The high resolution structure of allophycocyanin from cyanobacterium Nostoc sp. WR13
To Be Published
|
|
