7Z5X
 
 | | ROS1 with AstraZeneca ligand 2 | | Descriptor: | (2R)-2-[5-(6-amino-5-{(1R)-1-[2-(1,3-dihydro-2H-1,2,3-triazol-2-yl)-5-fluorophenyl]ethoxy}pyridin-3-yl)-4-methyl-1,3-thiazol-2-yl]propane-1,2-diol, Proto-oncogene tyrosine-protein kinase ROS | | Authors: | Hargreaves, D. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.035 Å) | | Cite: | Virtual Screening in the Cloud Identifies Potent and Selective ROS1 Kinase Inhibitors.
J.Chem.Inf.Model., 62, 2022
|
|
6Q3Z
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the inhibitor 16k | | Descriptor: | (7~{R})-2-[[2-ethoxy-4-(1-methylpiperidin-4-yl)phenyl]amino]-7-ethyl-5-methyl-8-[(4-methylthiophen-2-yl)methyl]-7~{H}-pteridin-6-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Heidenreich, D, Watts, E, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S, Hoelder, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designing Dual Inhibitors of Anaplastic Lymphoma Kinase (ALK) and Bromodomain-4 (BRD4) by Tuning Kinase Selectivity.
J.Med.Chem., 62, 2019
|
|
4F9L
 
 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the 20.1 Single Chain Antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 20.1 anti-BTN3A1 antibody fragment, Butyrophilin subfamily 3 member A1 | | Authors: | Palakodeti, A, Sandstrom, A, Sundaresan, L, Harly, C, Nedellec, S, Olive, D, Scotet, E, Bonneville, M, Adams, E.J. | | Deposit date: | 2012-05-18 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1404 Å) | | Cite: | The molecular basis for modulation of human V(gamma)9V(delta)2 T cell responses by CD277/Butyrophilin-3 (BTN3A)-specific antibodies
J.Biol.Chem., 287, 2012
|
|
1BAR
 
 | | THREE-DIMENSIONAL STRUCTURES OF ACIDIC AND BASIC FIBROBLAST GROWTH FACTORS | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Zhu, X, Komiya, H, Chirino, A, Faham, S, Fox, G.M, Arakawa, T, Hsu, B.T, Rees, D.C. | | Deposit date: | 1992-09-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structures of acidic and basic fibroblast growth factors.
Science, 251, 1991
|
|
7S4E
 
 | |
7KPA
 
 | | asymmetric hTNF-alpha | | Descriptor: | 5-(1-{[2-(difluoromethoxy)phenyl]methyl}-2-{[3-(2-oxopyrrolidin-1-yl)phenoxy]methyl}-1H-benzimidazol-6-yl)pyridin-2(1H)-one, Tumor necrosis factor | | Authors: | Fox III, D, Lukacs, C.M, Ceska, T. | | Deposit date: | 2020-11-10 | | Release date: | 2021-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A conformation-selective monoclonal antibody against a small molecule-stabilised signalling-deficient form of TNF.
Nat Commun, 12, 2021
|
|
4Y7I
 
 | | Crystal Structure of MTMR8 | | Descriptor: | Myotubularin-related protein 8, PHOSPHATE ION | | Authors: | Yoo, K, Lee, J, Son, J, Shin, W, Im, D, Heo, Y.S. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the catalytic phosphatase domain of MTMR8: implications for dimerization, membrane association and reversible oxidation.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1BFT
 
 | |
8HB1
 
 | | Crystal structure of NAD-II riboswitch (two strands) with NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, MAGNESIUM ION, RNA (30-MER), ... | | Authors: | Peng, X, Lilley, D.M.J, Huang, L. | | Deposit date: | 2022-10-27 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Nucleic Acids Res., 51, 2023
|
|
7Z2B
 
 | | P. berghei kinesin-8B motor domain in AMPPNP state bound to tubulin dimer | | Descriptor: | Detyrosinated tubulin alpha-1B chain, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-8, ... | | Authors: | Liu, T, Shilliday, F, Cook, A.D, Moores, C.A. | | Deposit date: | 2022-02-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanochemical tuning of a kinesin motor essential for malaria parasite transmission.
Nat Commun, 13, 2022
|
|
7VEU
 
 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with galacturonic acid | | Descriptor: | GLYCEROL, SPH1118, alpha-D-galactopyranuronic acid | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
6AOB
 
 | |
5KC4
 
 | | Structure of TmRibU, orthorhombic crystal form | | Descriptor: | RIBOFLAVIN, Riboflavin transporter RibU, nonyl beta-D-glucopyranoside | | Authors: | Karpowich, N.K, Wang, D.N, Song, J.M. | | Deposit date: | 2016-06-04 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An Aromatic Cap Seals the Substrate Binding Site in an ECF-Type S Subunit for Riboflavin.
J.Mol.Biol., 428, 2016
|
|
5JZ9
 
 | | Crystal structure of HsaD bound to 3,5-dichloro-4-hydroxybenzenesulphonic acid | | Descriptor: | 3,5-dichloro-4-hydroxybenzene-1-sulfonic acid, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase | | Authors: | Ryan, A, Polycarpou, E, Lack, N.A, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E, Ballet, R, Abihammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-16 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
7KEG
 
 | | Crystal structure from SARS-COV2 NendoU NSP15 | | Descriptor: | PHOSPHATE ION, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | Deposit date: | 2020-10-10 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7KFI
 
 | | SARS-CoV-2 Main protease immature form - apo structure | | Descriptor: | 3C-like proteinase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE | | Authors: | Noske, G.D, Nakamura, A.M, Gawriljuk, V.O, Lima, G.M.A, Zeri, A.C.M, Nascimento, A.F.Z, Oliva, G, Godoy, A.S. | | Deposit date: | 2020-10-14 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Crystallographic Snapshot of SARS-CoV-2 Main Protease Maturation Process.
J.Mol.Biol., 433, 2021
|
|
5MUZ
 
 | | Structure of a C-terminal domain of a reptarenavirus L protein | | Descriptor: | L protein | | Authors: | Rosenthal, M, Gogrefe, N, Reguera, J, Vogel, D, Rauschenberger, B, Cusack, S, Gunther, S, Reindl, S. | | Deposit date: | 2017-01-14 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.776 Å) | | Cite: | Structural insights into reptarenavirus cap-snatching machinery.
PLoS Pathog., 13, 2017
|
|
7Z5W
 
 | | ROS1 with AstraZeneca ligand 1 | | Descriptor: | Proto-oncogene tyrosine-protein kinase ROS, SULFATE ION, ~{N}-[6-methyl-2-[(2~{S})-2-[3-(3-methylpyrazin-2-yl)-1,2-oxazol-5-yl]pyrrolidin-1-yl]pyrimidin-4-yl]-1,3-thiazol-2-amine | | Authors: | Hargreaves, D. | | Deposit date: | 2022-03-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Virtual Screening in the Cloud Identifies Potent and Selective ROS1 Kinase Inhibitors.
J.Chem.Inf.Model., 62, 2022
|
|
8HB8
 
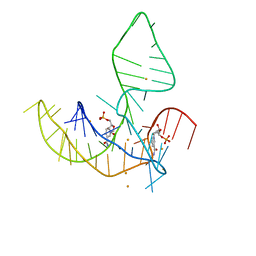 | |
3IDX
 
 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C222 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab b13 heavy chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
1BEV
 
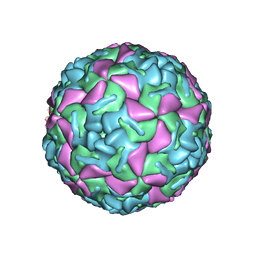 | | BOVINE ENTEROVIRUS VG-5-27 | | Descriptor: | BOVINE ENTEROVIRUS COAT PROTEINS VP1 TO VP4, MYRISTIC ACID, SULFATE ION | | Authors: | Smyth, M, Tate, J, Lyons, C, Hoey, E, Martin, S, Stuart, D. | | Deposit date: | 1996-04-03 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Implications for viral uncoating from the structure of bovine enterovirus.
Nat.Struct.Biol., 2, 1995
|
|
5Y6F
 
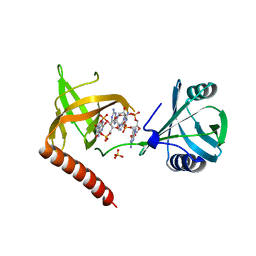 | | Crystal structure of YcgR in complex with c-di-GMP from Escherichia coli | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Flagellar brake protein YcgR, SULFATE ION | | Authors: | Hou, Y.J, Wang, D.C, Li, D.F. | | Deposit date: | 2017-08-11 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility inEscherichia coli.
J.Biol.Chem., 295, 2020
|
|
1BHN
 
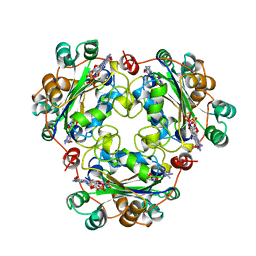 | | NUCLEOSIDE DIPHOSPHATE KINASE ISOFORM A FROM BOVINE RETINA | | Descriptor: | GUANOSINE-3',5'-MONOPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, NUCLEOSIDE DIPHOSPHATE TRANSFERASE | | Authors: | Ladner, J.E, Abdulaev, N.G, Kakuev, D.L, Karaschuk, G.N, Tordova, M, Eisenstein, E, Fujiwara, J.H, Ridge, K.D, Gilliland, G.L. | | Deposit date: | 1998-06-10 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The three-dimensional structures of two isoforms of nucleoside diphosphate kinase from bovine retina.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
4YFT
 
 | | HUab-20bp | | Descriptor: | DNA-binding protein HU-alpha, DNA-binding protein HU-beta, synthetic DNA strand | | Authors: | Hammel, M, Reyes, F.E, Parpana, R, Tainer, J.A, Adhya, S, Amlanjyoti, D. | | Deposit date: | 2015-02-25 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.914 Å) | | Cite: | HU multimerization shift controls nucleoid compaction.
Sci Adv, 2, 2016
|
|
6W9O
 
 | |
