2RI9
 
 | | Penicillium citrinum alpha-1,2-mannosidase in complex with a substrate analog | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lobsanov, Y.D, Yoshida, T, Desmet, T, Nerinckx, W, Yip, P, Claeyssens, M, Herscovics, A, Howell, P.L. | | Deposit date: | 2007-10-10 | | Release date: | 2008-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Modulation of activity by Arg407: structure of a fungal alpha-1,2-mannosidase in complex with a substrate analogue.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6HUK
 
 | | CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with bicuculline and megabody Mb38. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-1,Gamma-aminobutyric acid receptor subunit alpha-1, Gamma-aminobutyric acid receptor subunit beta-3, ... | | Authors: | Masiulis, S, Desai, R, Uchanski, T, Serna Martin, I, Laverty, D, Karia, D, Malinauskas, T, Jasenko, Z, Pardon, E, Kotecha, A, Steyaert, J, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2018-10-08 | | Release date: | 2019-01-02 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | GABAAreceptor signalling mechanisms revealed by structural pharmacology.
Nature, 565, 2019
|
|
7RUX
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-8311 | | Descriptor: | B-cell lymphoma 6 protein, FORMIC ACID, N-(3-chloropyridin-4-yl)-2-(3-(cyclopropylmethyl)-4-oxo-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)acetamide | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-08-18 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the BCL6 BTB domain in complex with OICR-8311
To Be Published
|
|
7RV0
 
 | |
7RV2
 
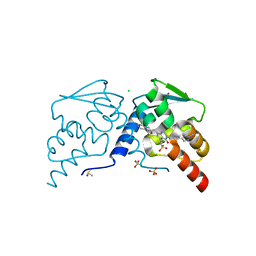 | |
7RUW
 
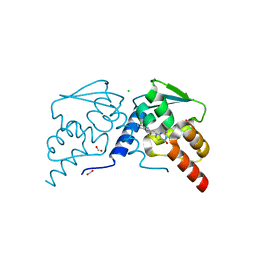 | | Crystal structure of the BCL6 BTB domain in complex with OICR-7859 | | Descriptor: | 2-(2-amino-4-oxo-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-N-(3-chloropyridin-4-yl)acetamide, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-08-18 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the BCL6 BTB domain in complex with OICR-7859
To Be Published
|
|
5UF6
 
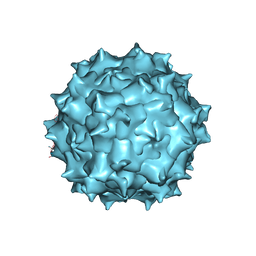 | | The 2.8 A Electron Microscopy Structure of Adeno-Associated Virus-DJ Bound by a Heparanoid Pentasaccharide | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, capsid protein VP1 | | Authors: | Xie, Q, Spear, J.M, Noble, A.J, Sousa, D.R, Meyer, N.L, Davulcu, O, Zhang, F, Linhardt, R.J, Stagg, S.M, Chapman, M. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The 2.8 angstrom Electron Microscopy Structure of Adeno-Associated Virus-DJ Bound by a Heparinoid Pentasaccharide.
Mol Ther Methods Clin Dev, 5, 2017
|
|
7K9I
 
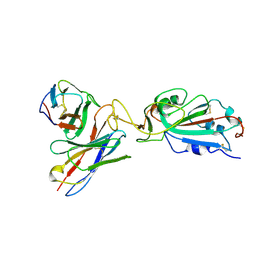 | |
6HJN
 
 | | Structure of Influenza Hemagglutinin ectodomain (A/duck/Alberta/35/76) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Benton, D.J, Rosenthal, P.B. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-26 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Influenza hemagglutinin membrane anchor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7RUZ
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-8445 | | Descriptor: | 2-[2-(benzylamino)-4-oxo-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl]-N-(3-chloropyridin-4-yl)acetamide, B-cell lymphoma 6 protein, CHLORIDE ION, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-08-18 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of the BCL6 BTB domain in complex with OICR-8445
To Be Published
|
|
7RV1
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-8826 | | Descriptor: | (E)-3-(7-(2-((3-chloropyridin-4-yl)amino)-2-oxoethyl)-3-(3-(1-methyl-1H-pyrazol-4-yl)prop-2-yn-1-yl)-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)acrylamide, B-cell lymphoma 6 protein, DIMETHYL SULFOXIDE, ... | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-08-18 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystal structure of the BCL6 BTB domain in complex with OICR-8826
To Be Published
|
|
6V0K
 
 | |
8OF3
 
 | | Structure of the apoform of ALDEHYDE DEHYDROGENASE 5F1 (ALDH5F1) from the moss Physcomitrium patens | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Morera, S, Kopecny, D, Vigouroux, A. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.348 Å) | | Cite: | A study on abiotic stress responses of aldehyde dehydrogenase (ALDH) superfamilies in moss and barley focused on members linked to the GABA shunt pathway
To Be Published
|
|
7RUY
 
 | | Crystal structure of the BCL6 BTB domain in complex with OICR-8388 | | Descriptor: | B-cell lymphoma 6 protein, FORMIC ACID, N-(3-chloropyridin-4-yl)-2-(2-{[(2,4-dimethoxyphenyl)methyl]amino}-4-oxo-3,4-dihydro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)acetamide | | Authors: | Kuntz, D.A, Prive, G.G. | | Deposit date: | 2021-08-18 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of the BCL6 BTB domain in complex with OICR-8388
To Be Published
|
|
6XQ6
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragment 11 | | Descriptor: | 3-phenoxyphenol, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6XQ3
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-(((3-(4-Carboxyphenoxy)benzyl)oxy)methyl)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 3-(3-{[3-(4-carboxyphenoxy)phenyl]methoxy}phenyl)-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6XQ8
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragments 1 & 11 | | Descriptor: | 3-phenoxyphenol, 7-methyl-3-phenyl-1H-indole-2-carboxylic acid, ACETATE ION, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6RUT
 
 | |
6XQ5
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragment 1 | | Descriptor: | 7-methyl-3-phenyl-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6XQ7
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragment 5 | | Descriptor: | 5-bromo-3-methyl-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6EZC
 
 | | Crystal Structure of human tRNA-dihydrouridine(20) synthase catalytic domain E294K Q305K double mutant | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Bou-Nader, C, Bregeon, D, Vincent, G, Fontecave, M, Hamdane, D. | | Deposit date: | 2017-11-14 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Electrostatic Potential in the tRNA Binding Evolution of Dihydrouridine Synthases.
Biochemistry, 57, 2018
|
|
8T3L
 
 | | TRPV1 in nanodisc bound with 2 LPA molecules in neighboring monomers | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, (2R)-3-{[(R)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctadecanoate, SODIUM ION, ... | | Authors: | Arnold, W.R, Julius, D, Cheng, Y. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of TRPV1 modulation by endogenous bioactive lipids.
Nat.Struct.Mol.Biol., 2024
|
|
8OJ1
 
 | |
3HK2
 
 | | Crystal structure of T. thermophilus Argonaute N478 mutant protein complexed with DNA guide strand and 19-nt RNA target strand | | Descriptor: | 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*AP*GP*T)-3', 5'-R(*UP*AP*UP*AP*CP*AP*AP*CP*CP*UP*AP*CP*UP*AP*CP*CP*UP*CP*G)-3', Argonaute, ... | | Authors: | Wang, Y, Li, H, Sheng, G, Patel, D.J. | | Deposit date: | 2009-05-22 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes.
Nature, 461, 2009
|
|
6XOD
 
 | | Crystal structure of the PEX4-PEX22 protein complex from Arabidopsis thaliana | | Descriptor: | Peroxisome biogenesis protein 22, Protein PEROXIN-4 | | Authors: | Olmos Jr, J.L, Bradford, S.E, Miller, M.D, Xu, W, Wright, Z.J, Bartel, B, Phillips Jr, G.N. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Structure of the Arabidopsis PEX4-PEX22 Peroxin Complex-Insights Into Ubiquitination at the Peroxisomal Membrane
Front Cell Dev Biol, 10, 2022
|
|
