6TE9
 
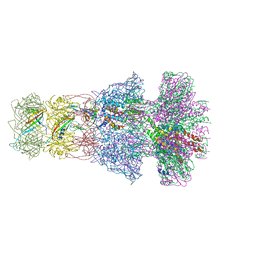 | | Neck of native GTA particle computed with C6 symmetry | | Descriptor: | Adaptor protein Rcc01688, Phage major tail protein, TP901-1 family, ... | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-11-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
7S24
 
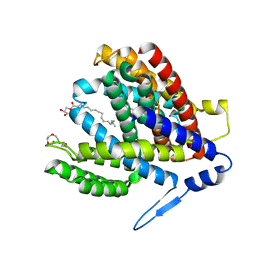 | | Crystal structure of the Na+/H+ antiporter NhaA at pH 6.5 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Na(+)/H(+) antiporter NhaA, PENTAETHYLENE GLYCOL | | Authors: | Drew, D, Brock, J, Uzdavinys, P, Matsuoka, R. | | Deposit date: | 2021-09-03 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Na + /H + antiporter NhaA at active pH reveals the mechanistic basis for pH sensing.
Nat Commun, 13, 2022
|
|
7RMN
 
 | |
1IXR
 
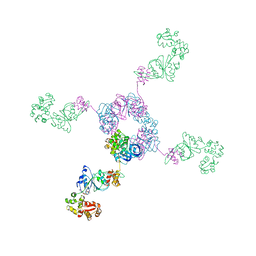 | | RuvA-RuvB complex | | Descriptor: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | Authors: | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
4UAG
 
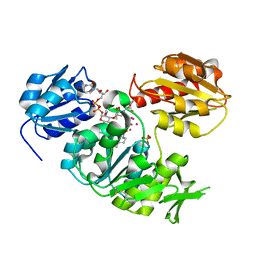 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | SULFATE ION, UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, UNKNOWN ATOM OR ION, ... | | Authors: | Bertrand, J, Auger, G, Martin, L, Fanchon, E, Blanot, D, Le Beller, D, Van Heijenoort, J, Dideberg, O. | | Deposit date: | 1999-03-09 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Determination of the MurD mechanism through crystallographic analysis of enzyme complexes.
J.Mol.Biol., 289, 1999
|
|
7RXF
 
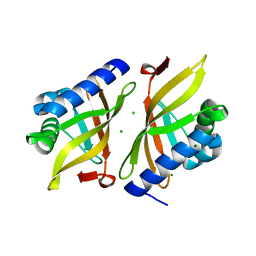 | |
7RZN
 
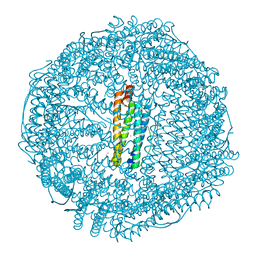 | | Crystal Structure of Ferritin grown by microbatch method in presence of agarose and electric field 2.1KV | | Descriptor: | CADMIUM ION, Ferritin light chain, SULFATE ION | | Authors: | Aditya, S, Priyadharshine, R, Maham, I, Miller, J.D, Stojanoff, V. | | Deposit date: | 2021-08-27 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of Ferritin grown by microbatch method in presence of agarose and electric field 2.1KV
To Be Published
|
|
5R62
 
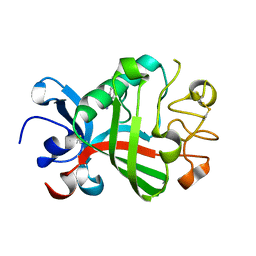 | | PanDDA analysis group deposition -- Crystal Structure of FIBRINOGEN-LIKE GLOBE DOMAIN OF HUMAN TENASCIN-C in complex with Z2856434840 | | Descriptor: | 4-(4-methylpiperazin-1-yl)benzonitrile, Tenascin C (Hexabrachion), isoform CRA_a | | Authors: | Coker, J.A, Bezerra, G.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Yue, W.W, Marsden, B.D. | | Deposit date: | 2020-02-28 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3HIZ
 
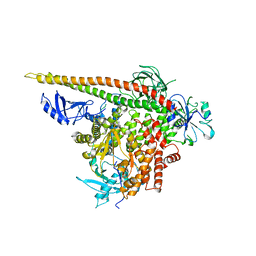 | | Crystal structure of p110alpha H1047R mutant in complex with niSH2 of p85alpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Amzel, L.M, Vogelstein, B, Gabelli, S.B, Mandelker, D. | | Deposit date: | 2009-05-20 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A frequent kinase domain mutation that changes the interaction between PI3K{alpha} and the membrane.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7RYW
 
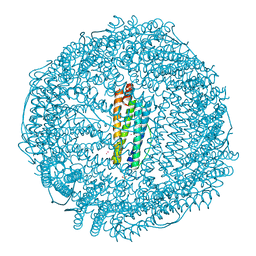 | | Crystal structure of Ferritin grown by the microbatch method in the presence of Agarose | | Descriptor: | CADMIUM ION, Ferritin light chain, SULFATE ION | | Authors: | Aditya, S, Priyadharshine, R, Maham, I, Miller, D.J, Stojanoff, V. | | Deposit date: | 2021-08-26 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of Ferritin grown by microbatch method in the presence of agarose
To Be Published
|
|
6TPN
 
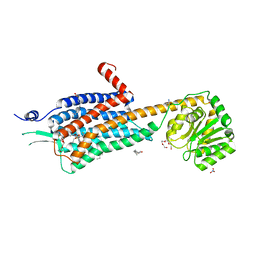 | | Crystal structure of the Orexin-2 receptor in complex with HTL6641 at 2.61 A resolution | | Descriptor: | 2-(5,6-dimethoxypyridin-3-yl)-1,1-bis(oxidanylidene)-4-[[2,4,6-tris(fluoranyl)phenyl]methyl]pyrido[2,3-e][1,2,4]thiadiazin-3-one, NITRATE ION, OLEIC ACID, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.608 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
7RZX
 
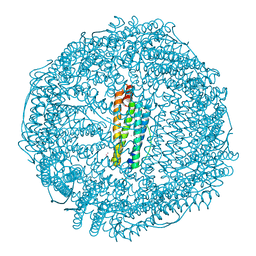 | | Crystal Structure of Ferritin grown by microbatch method in presence of agarose and electric field 4.3KV | | Descriptor: | CADMIUM ION, Ferritin light chain, SULFATE ION | | Authors: | Aditya, S, Priyadharshine, R, Maham, I, Miller, J.D, Stojanoff, V. | | Deposit date: | 2021-08-27 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structure of Ferritin grown by microbatch method in presence of agarose and electric field 4.3KV
To Be Published
|
|
7S3J
 
 | | Crystal Structure of AspB P450 in complex with brevianamide F substrates | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, AspB, GLYCEROL, ... | | Authors: | Newmister, S.A, Shende, V.V, Harris, N.R, Sanders, J.N, Khatri, Y, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2021-09-07 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular Dynamics Simulations Guide Chimeragenesis and Engineered Control of Chemoselectivity in Diketopiperazine Dimerases.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7SEH
 
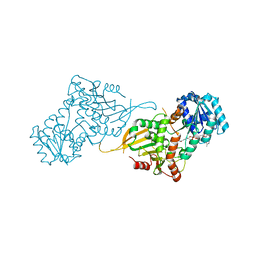 | | Glucose-6-phosphate 1-dehydrogenase (K403QdLtL) | | Descriptor: | Glucose-6-phosphate 1-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mathews, I.I, Garcia, A.A, Wakatsuki, S, Mochly-Rosen, D. | | Deposit date: | 2021-09-30 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Stabilization of glucose-6-phosphate dehydrogenase oligomers enhances catalytic activity and stability of clinical variants.
J.Biol.Chem., 298, 2022
|
|
6Q1J
 
 | |
3CBJ
 
 | | Chagasin-Cathepsin B complex | | Descriptor: | Cathepsin B, Chagasin, PHOSPHATE ION | | Authors: | Redzynia, I, Bujacz, G.D, Abrahamson, M, Ljunggren, A, Jaskolski, M, Mort, J.S. | | Deposit date: | 2008-02-22 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Displacement of the occluding loop by the parasite protein, chagasin, results in efficient inhibition of human cathepsin B.
J.Biol.Chem., 283, 2008
|
|
6Q5H
 
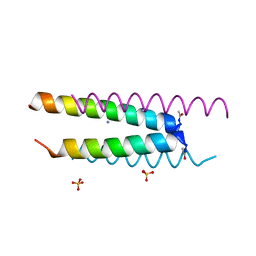 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24D | | Descriptor: | AMMONIUM ION, CC-Hex*-L24D, SULFATE ION | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
6Q5K
 
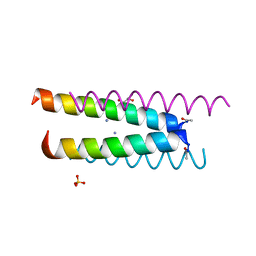 | | Crystal structure of a CC-Hex mutant that forms an antiparallel four-helix coiled coil CC-Hex*-L24K | | Descriptor: | AMMONIUM ION, CC-Hex*-L24K, GLYCEROL, ... | | Authors: | Wood, C.W, Beesley, J.L, Rhys, G.G, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2018-12-09 | | Release date: | 2019-05-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Navigating the Structural Landscape of De Novo alpha-Helical Bundles.
J.Am.Chem.Soc., 141, 2019
|
|
7F3X
 
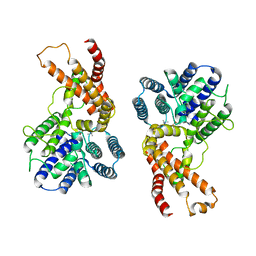 | | Lysophospholipid acyltransferase LPCAT3 in complex with lysophosphatidylcholine | | Descriptor: | LPCAT3, [2-((1-OXODODECANOXY-(2-HYDROXY-3-PROPANYL))-PHOSPHONATE-OXY)-ETHYL]-TRIMETHYLAMMONIUM | | Authors: | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Shen, Y, Cao, Y. | | Deposit date: | 2021-06-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
7RXK
 
 | |
7RY4
 
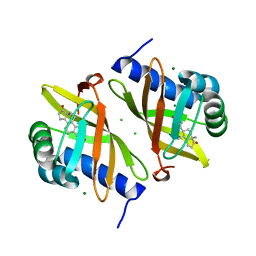 | | Multi-conformer model of Ketosteroid Isomerase Y57F/D40N mutant from Pseudomonas Putida (pKSI) bound to a transition state analog at 250 K | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Yabukarski, F, Doukov, T, Herschlag, D. | | Deposit date: | 2021-08-24 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Ensemble-function relationships to dissect mechanisms of enzyme catalysis.
Sci Adv, 8, 2022
|
|
3HHM
 
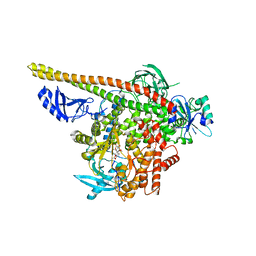 | | Crystal structure of p110alpha H1047R mutant in complex with niSH2 of p85alpha and the drug wortmannin | | Descriptor: | (1S,6BR,9AS,11R,11BR)-9A,11B-DIMETHYL-1-[(METHYLOXY)METHYL]-3,6,9-TRIOXO-1,6,6B,7,8,9,9A,10,11,11B-DECAHYDRO-3H-FURO[4, 3,2-DE]INDENO[4,5-H][2]BENZOPYRAN-11-YL ACETATE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, ... | | Authors: | Amzel, L.M, Vogelstein, B, Gabelli, S.B, Mandelker, D. | | Deposit date: | 2009-05-15 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A frequent kinase domain mutation that changes the interaction between PI3K{alpha} and the membrane.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3Q5J
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin | | Descriptor: | (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-19-{[3-(dimethylamino)propyl]amino}-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, Heat shock protein 83-1 | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin.
To be Published
|
|
1LV4
 
 | | Human catestatin 21-mer | | Descriptor: | catestatin | | Authors: | O'Connor, D.T, Preece, N.E. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational preferences and activities of peptides from the catecholamine release-inhibitory (catestatin) region of chromogranin A.
Regul.Pept., 118, 2004
|
|
5I68
 
 | | Alcohol oxidase from Pichia pastoris | | Descriptor: | Alcohol oxidase 1, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Vonck, J, Mills, D.J, Parcej, D.N. | | Deposit date: | 2016-02-16 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure of Alcohol Oxidase from Pichia pastoris by Cryo-Electron Microscopy.
Plos One, 11, 2016
|
|
