8VIJ
 
 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin Y34F C51S C71S variant (cyanomet) | | Descriptor: | CYANIDE ION, Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lecomte, J.T.J, Schlessman, J.L, Martinez, J.E, Schultz, T.D, Siegler, M.A, DelCampo, M, Le Magueres, P. | | Deposit date: | 2024-01-04 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin Y34F C51S C71S variant in the cyanomet state
To be published
|
|
6J60
 
 | | hnRNP A1 reversible amyloid core GFGGNDNFG (residues 209-217) | | Descriptor: | 9-mer peptide (GFGGNDNFG) from Heterogeneous nuclear ribonucleoprotein A1 | | Authors: | Luo, F, Zhou, H, Gui, X, Li, D, Li, X, Liu, C. | | Deposit date: | 2019-01-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.96 Å) | | Cite: | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
6N3K
 
 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 1 | | Descriptor: | Epoxide hydrolase, N-{cis-4-[(2,6-difluorophenyl)methoxy]cyclohexyl}-N'-(3-phenylpropyl)urea | | Authors: | de Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
6IWM
 
 | |
6N86
 
 | |
8C0B
 
 | | CryoEM structure of Aspergillus nidulans UTP-glucose-1-phosphate uridylyltransferase | | Descriptor: | UTP--glucose-1-phosphate uridylyltransferase | | Authors: | Han, X, D Angelo, C, Otamendi, A, Cifuente, J.O, de Astigarraga, E, Ochoa-Lizarralde, B, Grininger, M, Routier, F.H, Guerin, M.E, Fuehring, J, Etxebeste, O, Connell, S.R. | | Deposit date: | 2022-12-16 | | Release date: | 2023-06-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | CryoEM analysis of the essential native UDP-glucose pyrophosphorylase from Aspergillus nidulans reveals key conformations for activity regulation and function.
Mbio, 14, 2023
|
|
6JAF
 
 | | Crystal structure of Trypanosoma brucei gambiense glycerol kinase complex with PPi (pyrophosphatase reaction) | | Descriptor: | GLYCEROL, Glycerol kinase, PYROPHOSPHATE 2- | | Authors: | Balogun, E.O, Chishima, T, Ichinose, M, Inaoka, D.K, Kido, Y, Ibrahim, B, Bringaud, F, de Koning, H, McKerrow, J.H, Watanabe, Y, Nozaki, T, Michels, P.A.M, Harada, S, Kita, K, Shiba, T. | | Deposit date: | 2019-01-24 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Glycerol Kinase of African Human Trypanosomes Possesses a Pyrophosphatase Activity.
To Be Published
|
|
4PSY
 
 | | 100K crystal structure of Escherichia coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Wilson, M.A, Wan, Q, Bennet, B.C, Dealwis, C, Ringe, D, Petsko, G.A. | | Deposit date: | 2014-03-08 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Toward resolving the catalytic mechanism of dihydrofolate reductase using neutron and ultrahigh-resolution X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 22, 2014
|
|
6J0K
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A3 mutant in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A3 | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
8CA2
 
 | |
3L0X
 
 | |
4PDW
 
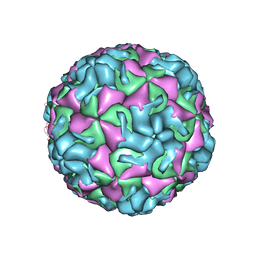 | | A benzonitrile analogue inhibits rhinovirus replication | | Descriptor: | 4-[(4,5-dimethoxy-2-nitrophenyl)acetyl]benzonitrile, Capsid protein VP4/VP2, GLYCEROL, ... | | Authors: | Querol-Audi, J, Guerra, P, Lacroix, C, Roche, M, Franco, D, Froeyen, M, Terme, T, Vanelle, P, Neyts, J, Leyssen, P, Verdaguer, N. | | Deposit date: | 2014-04-22 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel benzonitrile analogue inhibits rhinovirus replication.
J.Antimicrob.Chemother., 69, 2014
|
|
7SFB
 
 | |
7SF3
 
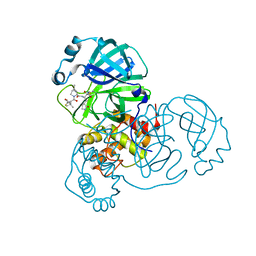 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML1006m | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-3-hydroxy-4-(methylamino)-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-02 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
3AGQ
 
 | | Structure of viral polymerase form II | | Descriptor: | Elongation factor Ts, Elongation factor Tu 1, LINKER, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2010-04-06 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Assembly of Q{beta} viral RNA polymerase with host translational elongation factors EF-Tu and -Ts
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7SFI
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML104 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[N-(2,4,6-trifluorophenyl)glycyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CALCIUM ION, ... | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-03 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational design of a new class of protease inhibitors for the potential treatment of coronavirus diseases
To Be Published
|
|
3N0K
 
 | | Proteinase inhibitor from Coprinopsis cinerea | | Descriptor: | Serine protease inhibitor 1 | | Authors: | Renko, M, Sabotic, J, Bleuler-Martinez, S, Kallert, S, Avanzo, P, Kos, J, Aebi, M, Kuenzler, M, Turk, D. | | Deposit date: | 2010-05-14 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Trypsin Inhibition and Entomotoxicity of Cospin, Serine Protease Inhibitor Involved in Defense of Coprinopsis cinerea Fruiting Bodies.
J.Biol.Chem., 287, 2012
|
|
7SFH
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML102 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-(3-phenylpropanoyl)-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CALCIUM ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-03 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rational design of a new class of protease inhibitors for the potential treatment of coronavirus diseases
To Be Published
|
|
8C6D
 
 | | Production of antigenically stable enterovirus A71 virus-like particles in Pichia pastoris as a vaccine candidate. | | Descriptor: | (2S,3R,4E)-2-aminooctadec-4-ene-1,3-diol, Genome polyprotein, Genome polyprotein (Fragment) | | Authors: | Kingston, N.J, Snowden, J.S, Stonehouse, N.J, Rowlands, D.J, Hogle, J.M. | | Deposit date: | 2023-01-11 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Production of antigenically stable enterovirus A71 virus-like particles in Pichia pastoris as a vaccine candidate.
Biorxiv, 2023
|
|
8CKS
 
 | | Crystal structure of Human Serum Albumin in complex with FESAN | | Descriptor: | 3,3'-commo-bis(1,2-dicarba-3-ferra-closo-dodecaborane), DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Dolot, R.M, Kaniowski, D, Ebenryter-Olbinska, K, Szczupak, P, Suwara, J, Nawrot, B.C. | | Deposit date: | 2023-02-16 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Human Serum Albumin in complex with FESAN
To Be Published
|
|
7SET
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML1000 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2021-10-01 | | Release date: | 2022-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
6UVS
 
 | |
3G7C
 
 | | Structure of the Phosphorylation Mimetic of Occludin C-term Tail | | Descriptor: | Occludin | | Authors: | Tash, B.R, Sundstrom, J.M, Murakami, T, Flanagan, J.M, Bewley, M.C, Antonetii, D.A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and analysis of occludin phosphosites: a combined mass spectrometry and bioinformatics approach.
J.PROTEOME RES., 8, 2009
|
|
8C0Q
 
 | | Crystal structure of human carbonic anhydrase II in complex with a coumarin derivative. | | Descriptor: | (4-methyl-2-oxidanylidene-chromen-7-yl)methanesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A Combined in Silico and Structural Study Opens New Perspectives on Aliphatic Sulfonamides, a Still Poorly Investigated Class of CA Inhibitors.
Biology (Basel), 12, 2023
|
|
6VU5
 
 | | Structure of G-alpha-q bound to its chaperone Ric-8A | | Descriptor: | Guanine nucleotide-binding protein G(q) subunit alpha, Resistance to inhibitors of cholinesterase-8A (Ric-8A) | | Authors: | Seven, A.B, Hilger, D. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of G alpha Proteins in Complex with Their Chaperone Reveal Quality Control Mechanisms.
Cell Rep, 30, 2020
|
|
