2EVZ
 
 | |
3E3K
 
 | | Structural characterization of a putative endogenous metal chelator in the periplasmic nickel transporter NikA (butane-1,2,4-tricarboxylate without nickel form) | | Descriptor: | (2R)-butane-1,2,4-tricarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Cherrier, M.V, Cavazza, C, Bochot, C, Lemaire, D, Fontecilla-Camps, J.C. | | Deposit date: | 2008-08-07 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of a putative endogenous metal chelator in the periplasmic nickel transporter NikA
Biochemistry, 47, 2008
|
|
3E48
 
 | | Crystal structure of a nucleoside-diphosphate-sugar epimerase (SAV0421) from Staphylococcus aureus, Northeast Structural Genomics Consortium Target ZR319 | | Descriptor: | MAGNESIUM ION, Putative nucleoside-diphosphate-sugar epimerase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Mao, L, Janjua, H, Xiao, R, Ciccosanti, C, Foote, E.L, Wang, D, Tong, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-08-11 | | Release date: | 2008-08-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: |
|
|
8JQU
 
 | | Crystal structure of GppNHp bound GTPase domain of Rab5a from Leishmania donovani | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Pandey, D, Zohib, M, Pal, R.K, Biswal, B.K, Arora, A. | | Deposit date: | 2023-06-14 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Crystal structure of GppNHp bound GTPase domain of Rab5a from Leishmania donovani
To Be Published
|
|
8JT4
 
 | |
5VKZ
 
 | | Crystal structure of Mdm12 and combinatorial reconstitution of Mdm12/Mmm1 ERMES complexes for structural studies | | Descriptor: | Mitochondrial distribution and morphology protein 12 | | Authors: | Egea, P.F, AhYoung, A.P, Lu, B, Tan, H.R, Cascio, D. | | Deposit date: | 2017-04-24 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal structure of Mdm12 and combinatorial reconstitution of Mdm12/Mmm1 ERMES complexes for structural studies.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
8JSD
 
 | | Alginate lyase mutant-D180G | | Descriptor: | Alginate lyase | | Authors: | Zhang, X, Pan, L.X, Yang, D.F. | | Deposit date: | 2023-06-19 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.414 Å) | | Cite: | Substrate-binding mode guided protein design of alginate lyase FlAlyA for altered end-product distribution
To Be Published
|
|
8C0V
 
 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in single seam state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
8C0W
 
 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in twin seam state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
7AJU
 
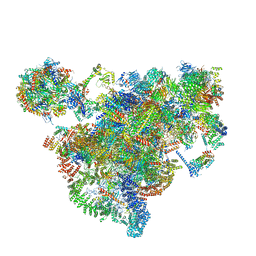 | | Cryo-EM structure of the 90S-exosome super-complex (state Post-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
7DAV
 
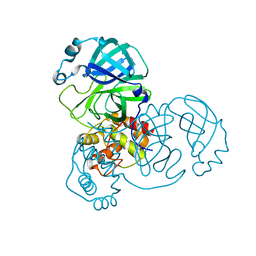 | | The native crystal structure of COVID-19 main protease | | Descriptor: | COVID-19 MAIN PROTEASE | | Authors: | He, Z.S, He, B, Cao, P, Jiang, H.D, Gong, Y, Gao, X.Y. | | Deposit date: | 2020-10-18 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A comparison of Remdesivir versus gold cluster in COVID-19 animal model: A better therapeutic outcome of gold cluster.
Nano Today, 44, 2022
|
|
8QZ5
 
 | |
8QYZ
 
 | |
5VHU
 
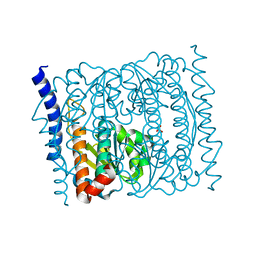 | | E. coli CFT073 c3406 | | Descriptor: | Isomerase, SULFATE ION | | Authors: | Cech, D.L, Pratt, A.C, Woodard, R.W. | | Deposit date: | 2017-04-13 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of arabinose-5-phosphate isomerases.
To Be Published
|
|
157D
 
 | | CRYSTAL AND MOLECULAR STRUCTURE OF R(CGCGAAUUAGCG): AN RNA DUPLEX CONTAINING TWO G(ANTI).A(ANTI) BASE-PAIRS | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*AP*GP*CP*G)-3') | | Authors: | Leonard, G.A, McAuley-Hecht, K.E, Ebel, S, Lough, D.M, Brown, T, Hunter, W.N. | | Deposit date: | 1994-02-01 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal and molecular structure of r(CGCGAAUUAGCG): an RNA duplex containing two G(anti).A(anti) base pairs.
Structure, 2, 1994
|
|
7AJT
 
 | | Cryo-EM structure of the 90S-exosome super-complex (state Pre-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
3EI6
 
 | | Crystal structure of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with PLP-DAP: an external aldimine mimic | | Descriptor: | (2S,6S)-2-amino-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]heptanedioic acid, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
8SV1
 
 | | Caspase-1 complex with interleukin-18 | | Descriptor: | Caspase-1, Interleukin-18 | | Authors: | Dong, Y, Pascal, D, Jon, K, Wu, H. | | Deposit date: | 2023-05-15 | | Release date: | 2024-05-08 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural transitions enable interleukin-18 maturation and signaling.
Immunity, 57, 2024
|
|
3ELZ
 
 | | Crystal structure of Zebrafish Ileal Bile Acid-Bindin Protein complexed with cholic acid (crystal form A). | | Descriptor: | CHOLIC ACID, ileal Bile Acid-Binding Protein | | Authors: | Capaldi, S, Saccomani, G, Fessas, D, Signorelli, M, Perduca, M, Monaco, H.L. | | Deposit date: | 2008-09-23 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The X-Ray structure of zebrafish (Danio rerio) ileal bile acid-binding protein reveals the presence of binding sites on the surface of the protein molecule.
J.Mol.Biol., 385, 2009
|
|
8C7C
 
 | | Double mutant V(M84)C/A(L278)C structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | Descriptor: | 1,2-ETHANEDIOL, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Gabdulkhakov, A, Selikhanov, G, Fufina, T, Vasilieva, L, Atamas, A, Uhimchuk, D. | | Deposit date: | 2023-01-14 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds.
Membranes (Basel), 13, 2023
|
|
8C6K
 
 | | Double mutant A(L53)C/I(L64)C structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A, Selikhanov, G, Fufina, T, Vasilieva, L, Atamas, A, Uhimchuk, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds.
Membranes (Basel), 13, 2023
|
|
7LRN
 
 | |
3EF1
 
 | |
8C5X
 
 | | Double mutant A(L37)C/S(L99)C structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A, Selikhanov, G, Fufina, T, Vasilieva, L, Atamas, A, Uhimchuk, D. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds.
Membranes (Basel), 13, 2023
|
|
8R5J
 
 | | Crystal structure of MERS-CoV main protease | | Descriptor: | Non-structural protein 11 | | Authors: | Balcomb, B.H, Fairhead, M, Koekemoer, L, Lithgo, R.M, Aschenbrenner, J.C, Chandran, A.V, Godoy, A.S, Lukacik, P, Marples, P.G, Mazzorana, M, Ni, X, Strain-Damerell, C, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-11-16 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of MERS-CoV main protease
To Be Published
|
|
