6ZKC
 
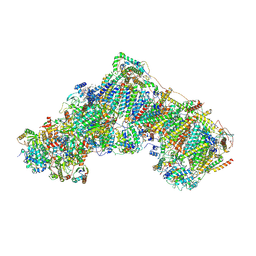 | | Complex I during turnover, closed | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
6ZKR
 
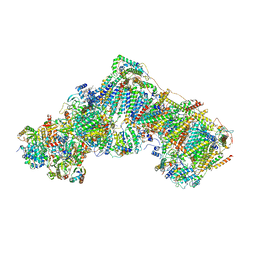 | | Native complex I, open3 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kampjut, D, Sazanov, L.A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The coupling mechanism of mammalian respiratory complex I.
Science, 370, 2020
|
|
5J5R
 
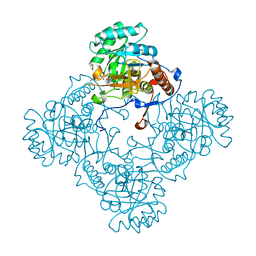 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor VCC234718 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, cyclohexyl{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}methanone | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-04-03 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Inosine Monophosphate Dehydrogenase, GuaB2, Is a Vulnerable New Bactericidal Drug Target for Tuberculosis.
ACS Infect Dis, 3, 2017
|
|
4UUN
 
 | |
5J37
 
 | | Crystal structure of 60-mer BFDV Capsid Protein in complex with single stranded DNA | | Descriptor: | Beak and feather disease virus capsid protein, PHOSPHATE ION, single stranded DNA | | Authors: | Sarker, S, Raidal, S, Aragao, D, Forwood, J.K. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the assembly and regulation of distinct viral capsid complexes.
Nat Commun, 7, 2016
|
|
4V7S
 
 | | Crystal structure of the E. coli ribosome bound to telithromycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2547 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1CJX
 
 | | CRYSTAL STRUCTURE OF PSEUDOMONAS FLUORESCENS HPPD | | Descriptor: | 4-HYDROXYPHENYLPYRUVATE DIOXYGENASE, ACETATE ION, ETHYL MERCURY ION, ... | | Authors: | Serre, L, Sailland, A, Sy, D, Boudec, P, Rolland, A, Pebay-Peroulla, E, Cohen-Addad, C. | | Deposit date: | 1999-04-20 | | Release date: | 2000-04-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Pseudomonas fluorescens 4-hydroxyphenylpyruvate dioxygenase: an enzyme involved in the tyrosine degradation pathway.
Structure Fold.Des., 7, 1999
|
|
4V0V
 
 | | The crystal structure of mouse PP1G in complex with truncated human PPP1R15B (631-660) | | Descriptor: | MANGANESE (II) ION, PROTEIN PHOSPHATASE 1 REGULATORY SUBUNIT 15B, SERINE/THREONINE-PROTEIN PHOSPHATASE PP1-GAMMA CATALYTIC SUBUNIT, ... | | Authors: | Chen, R, Yan, Y, Casado, A.C, Ron, D, Read, R.J. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | G-actin provides substrate-specificity to eukaryotic initiation factor 2 alpha holophosphatases.
Elife, 4, 2015
|
|
5OQS
 
 | | Solution structure of antifungal protein NFAP | | Descriptor: | NFAP | | Authors: | Hajdu, D, Czajlik, A, Marx, F, Galgoczy, L, Batta, G. | | Deposit date: | 2017-08-14 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and novel insights into phylogeny and mode of action of the Neosartorya (Aspergillus) fischeri antifungal protein (NFAP).
Int.J.Biol.Macromol., 129, 2019
|
|
6ZD2
 
 | | Structure of apo telomerase from Candida Tropicalis truncated from C-terminal domain | | Descriptor: | SODIUM ION, Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-13 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
6ZDQ
 
 | | Structure of telomerase from Candida albicans in complexe with TWJ fragment of telomeric RNA | | Descriptor: | RNA, Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-15 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
6ZD1
 
 | | Structure of apo telomerase from Candida Tropicalis | | Descriptor: | Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-13 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
6ZD6
 
 | | Structure of apo telomerase from Candida Tropicalis | | Descriptor: | Telomerase reverse transcriptase | | Authors: | Zhai, L, Rety, S, Chen, W.F, Auguin, D, Xi, X.G. | | Deposit date: | 2020-06-13 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structures of N-terminally truncated telomerase reverse transcriptase from fungi‡.
Nucleic Acids Res., 49, 2021
|
|
8C0O
 
 | |
6YWF
 
 | |
8CNX
 
 | | Structure of Enterovirus D68 3C protease | | Descriptor: | Protease 3C | | Authors: | Lithgo, R.M, Fairhead, M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Godoy, A.S, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-02-24 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of EV D68 3C protease
To Be Published
|
|
1KS9
 
 | | Ketopantoate Reductase from Escherichia coli | | Descriptor: | 2-DEHYDROPANTOATE 2-REDUCTASE | | Authors: | Matak-Vinkovic, D, Vinkovic, M, Saldanha, S.A, Ashurst, J.A, von Delft, F, Inoue, T, Miguel, R.N, Smith, A.G, Blundell, T.L, Abell, C. | | Deposit date: | 2002-01-11 | | Release date: | 2002-01-25 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Escherichia coli ketopantoate reductase at 1.7 A resolution and insight into the enzyme mechanism.
Biochemistry, 40, 2001
|
|
1KSL
 
 | | STRUCTURE OF RSUA | | Descriptor: | RIBOSOMAL SMALL SUBUNIT PSEUDOURIDINE SYNTHASE A, URACIL | | Authors: | Sivaraman, J, Sauve, V, Larocque, R, Stura, E.A, Schrag, J.D, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-01-13 | | Release date: | 2002-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the 16S rRNA pseudouridine synthase RsuA bound to uracil and UMP.
Nat.Struct.Biol., 9, 2002
|
|
7SZ6
 
 | | Kinetically trapped Pseudomonas-phage PaP3 portal protein - delta barrel mutant class-3 | | Descriptor: | Portal protein | | Authors: | Hou, C.-F.D, Swanson, N.A, Li, F, Yang, R, Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2021-11-25 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (6.24 Å) | | Cite: | Cryo-EM Structure of a Kinetically Trapped Dodecameric Portal Protein from the Pseudomonas-phage PaP3.
J.Mol.Biol., 434, 2022
|
|
6E5V
 
 | | human mGlu8 receptor amino terminal domain in complex with (S)-3,4-Dicarboxyphenylglycine (DCPG) | | Descriptor: | 4-[(S)-amino(carboxy)methyl]benzene-1,2-dicarboxylic acid, CHLORIDE ION, Metabotropic glutamate receptor 8 | | Authors: | Chen, Q, Ho, J.D, Ashok, S, Vargas, M.C, Wang, J, Atwell, S, Bures, M, Schkeryantz, J.M, Monn, J.A, Hao, J. | | Deposit date: | 2018-07-23 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis for ( S)-3,4-Dicarboxyphenylglycine (DCPG) As a Potent and Subtype Selective Agonist of the mGlu8Receptor.
J. Med. Chem., 61, 2018
|
|
5J7S
 
 | | Crystal structure of SM1-71 bound to TAK1-TAB1 | | Descriptor: | Mitogen-activated protein kinase kinase kinase 7/TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera, N-{2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]phenyl}propanamide | | Authors: | Gurbani, D, Westover, K.D. | | Deposit date: | 2016-04-06 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | Structure-guided development of covalent TAK1 inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
6E9C
 
 | | Selenomethionine Derivative Structure of A Bacterial Homolog to Human Lysosomal Transporter, Spinster | | Descriptor: | Major facilitator family transporter | | Authors: | Zhou, F, Yao, D, Rao, B, Zhang, L, Cao, Y. | | Deposit date: | 2018-07-31 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a bacterial homolog to human lysosomal transporter, spinster
Sci Bull (Beijing), 64, 2019
|
|
6E9T
 
 | | DHF58 filament | | Descriptor: | DHF58 filament | | Authors: | Lynch, E.M, Shen, H, Fallas, J.A, Kollman, J.M, Baker, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | De novo design of self-assembling helical protein filaments.
Science, 362, 2018
|
|
6ECZ
 
 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | Carbonic anhydrase 2, N-[2-hydroxy-3-nitro-5-(nitrosulfonyl)phenyl]-N'-(pentafluorophenyl)urea, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-08 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
6E84
 
 | | Crystal structure of the Corn aptamer in complex with TO | | Descriptor: | 1-methyl-4-[(Z)-(3-methyl-1,3-benzothiazol-2(3H)-ylidene)methyl]quinolin-1-ium, POTASSIUM ION, RNA (36-MER) | | Authors: | Sjekloca, L, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-27 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Binding between G Quadruplexes at the Homodimer Interface of the Corn RNA Aptamer Strongly Activates Thioflavin T Fluorescence.
Cell Chem Biol, 26, 2019
|
|
