1KNC
 
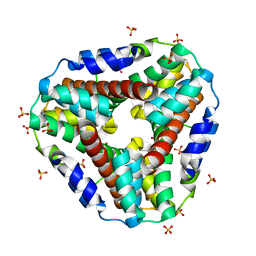 | | Structure of AhpD from Mycobacterium tuberculosis, a novel enzyme with thioredoxin-like activity. | | Descriptor: | AhpD protein, SULFATE ION | | Authors: | Bryk, R, Lima, C.D, Erdjument-Bromage, H, Tempst, P, Nathan, C. | | Deposit date: | 2001-12-18 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein.
Science, 295, 2002
|
|
6YX7
 
 | | The high resolution structure of allophycocyanin from cyanobacterium Nostoc sp. WR13, the P21212 crystal form. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Patel, H.M, Roszak, A.W, Madamwar, D, Cogdell, R.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | The high resolution structure of allophycocyanin from cyanobacterium Nostoc sp. WR13
To Be Published
|
|
8C6O
 
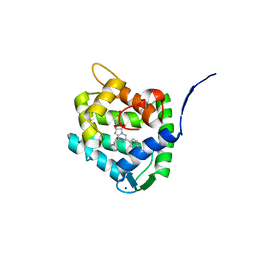 | | Crystal Structure of H64F obelin mutant from Obelia longissima at 2.2 Angstrom resolution | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, Obelin, SODIUM ION | | Authors: | Natashin, P.V, Burakova, L.P, Kovaleva, M.I, Schevtsov, M.B, Dmitrieva, D.A, Eremeeva, E.V, Markova, S.V, Mishin, A.V, Borshchevskiy, V.I, Vysotski, E.S. | | Deposit date: | 2023-01-12 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Role of Tyr-His-Trp Triad and Water Molecule Near the N1-Atom of 2-Hydroperoxycoelenterazine in Bioluminescence of Hydromedusan Photoproteins: Structural and Mutagenesis Study.
Int J Mol Sci, 24, 2023
|
|
8C0E
 
 | | The lipid linked oligosaccharide polymerase Wzy and its regulating co-polymerase Wzz form a complex in vivo and in vitro | | Descriptor: | ECA polysaccharide chain length modulation protein | | Authors: | Weckener, M, Woodward, L.S, Clarke, B.R, Liu, H, Ward, P.N, Le Bas, A, Bhella, D, Whitfield, C, Naismith, J.H. | | Deposit date: | 2022-12-16 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The lipid linked oligosaccharide polymerase Wzy and its regulating co-polymerase, Wzz, from enterobacterial common antigen biosynthesis form a complex.
Open Biology, 13, 2023
|
|
8C0C
 
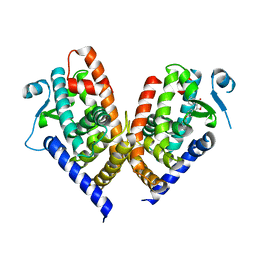 | | X-ray crystal structure of PPAR gamma ligand binding domain in complex with CZ46 | | Descriptor: | (2~{R})-2-[4-(naphthalen-1-ylmethoxy)phenyl]-4-oxidanyl-3-phenyl-2~{H}-furan-5-one, Peroxisome proliferator-activated receptor gamma | | Authors: | Capelli, D, Montanari, R, Pochetti, G, Villa, S, Meneghetti, F. | | Deposit date: | 2022-12-16 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biological Screening and Crystallographic Studies of Hydroxy gamma-Lactone Derivatives to Investigate PPAR gamma Phosphorylation Inhibition.
Biomolecules, 13, 2023
|
|
1KFZ
 
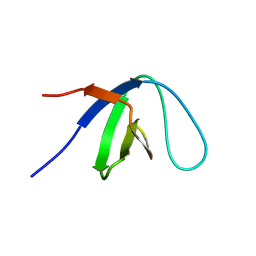 | | Solution Structure of C-terminal Sem-5 SH3 Domain (Ensemble of 16 Structures) | | Descriptor: | SEX MUSCLE ABNORMAL PROTEIN 5 | | Authors: | Ferreon, J.C, Volk, D.E, Luxon, B.A, Gorenstein, D, Hilser, V.J. | | Deposit date: | 2001-11-24 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Dynamics and Thermodynamics of the Native State Ensemble
of Sem-5 C-Terminal SH3 Domain
Biochemistry, 42, 2003
|
|
1KJ5
 
 | | Solution Structure of Human beta-defensin 1 | | Descriptor: | BETA-DEFENSIN 1 | | Authors: | Schibli, D.J, Hunter, H.N, Aseyev, V, Starner, T.D, Wiencek, J.M, McCray Jr, P.B, Tack, B.F, Vogel, H.J. | | Deposit date: | 2001-12-04 | | Release date: | 2002-03-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus.
J.Biol.Chem., 277, 2002
|
|
2OVI
 
 | |
7SSM
 
 | | Crystal structure of human STING R232 in complex with compound 11 | | Descriptor: | 2-({[(8R)-pyrazolo[1,5-a]pyrimidine-3-carbonyl]amino}methyl)-1-benzofuran-7-carboxylic acid, Stimulator of interferon genes protein | | Authors: | Sack, J.S, Critton, D.A. | | Deposit date: | 2021-11-11 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of Non-Nucleotide Small-Molecule STING Agonists via Chemotype Hybridization.
J.Med.Chem., 65, 2022
|
|
7SZL
 
 | | Crystal structure of the Toll/interleukin-1 receptor domain of human IL-1R10 (IL-1RAPL2) | | Descriptor: | X-linked interleukin-1 receptor accessory protein-like 2 | | Authors: | Nimma, S, Gu, W, Manik, M.K, Ve, T, Nanson, J.D, Kobe, B. | | Deposit date: | 2021-11-28 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Toll/interleukin-1 receptor (TIR) domain of IL-1R10 provides structural insights into TIR domain signalling.
Febs Lett., 596, 2022
|
|
8BVP
 
 | | Crystal structure of an N-terminal fragment of the effector protein Lpg2504 (SidI) from Legionella pneumophila | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Restriction endonuclease | | Authors: | Machtens, D.A, Willerding, J.M, Eschenburg, S, Reubold, T.F. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the N-terminal domain of the effector protein SidI of Legionella pneumophila reveals a glucosyl transferase domain.
Biochem.Biophys.Res.Commun., 661, 2023
|
|
1KG2
 
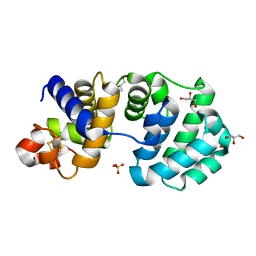 | | Crystal structure of the core fragment of MutY from E.coli at 1.2A resolution | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
1KG7
 
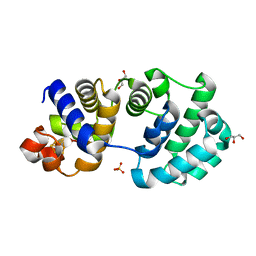 | | Crystal Structure of the E161A mutant of E.coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
8C0Z
 
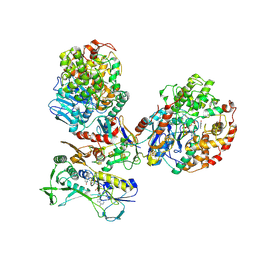 | | CryoEM structure of a tungsten-containing aldehyde oxidoreductase from Aromatoleum aromaticum | | Descriptor: | Aldehyde:ferredoxin oxidoreductase,tungsten-containing, BENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Winiarska, A, Ramirez-Amador, F, Hege, D, Gemmecker, Y, Prinz, S, Hochberg, G, Heider, J, Szaleniec, M, Schuller, J.M. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A bacterial tungsten-containing aldehyde oxidoreductase forms an enzymatic decorated protein nanowire.
Sci Adv, 9, 2023
|
|
1KT0
 
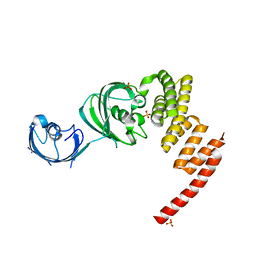 | | Structure of the Large FKBP-like Protein, FKBP51, Involved in Steroid Receptor Complexes | | Descriptor: | 51 KDA FK506-BINDING PROTEIN, SULFATE ION | | Authors: | Sinars, C.R, Cheung-Flynn, J, Rimerman, R.A, Scammell, J.G, Smith, D.F, Clardy, J.C. | | Deposit date: | 2002-01-14 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the large FK506-binding protein FKBP51, an Hsp90-binding protein and a
component of steroid receptor complexes
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7ZRU
 
 | |
1KH8
 
 | | Structure of a cis-proline (P114) to glycine variant of Ribonuclease A | | Descriptor: | CESIUM ION, SULFATE ION, pancreatic ribonuclease A | | Authors: | Schultz, D.A, Friedman, A.M, White, M.A, Fox, R.O. | | Deposit date: | 2001-11-29 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the cis-proline to glycine variant (P114G) of ribonuclease A.
Protein Sci., 14, 2005
|
|
2OU7
 
 | | Structure of the Catalytic Domain of Human Polo-like Kinase 1 | | Descriptor: | ACETATE ION, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Ding, Y.-H, Kothe, M, Kohls, D, Low, S. | | Deposit date: | 2007-02-09 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the catalytic domain of human polo-like kinase 1.
Biochemistry, 46, 2007
|
|
2P1D
 
 | |
8WB6
 
 | |
8WB7
 
 | |
5JYU
 
 | |
5JYT
 
 | |
7RZY
 
 | | CryoEM structure of Vibrio cholerae transposon Tn6677 AAA+ ATPase TnsC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Tn6677 Vibrio cholerae transposon TnsC (VchTnsC) | | Authors: | Hoffmann, F.T, Kim, M, Beh, L.Y, Wang, J, Vo, P.L.H, Gelsinger, D.R, Acree, C, Mohabir, J.T, Fernandez, I.S, Sternberg, S.H. | | Deposit date: | 2021-08-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Selective TnsC recruitment enhances the fidelity of RNA-guided transposition.
Nature, 609, 2022
|
|
2OMQ
 
 | |
