7ZUX
 
 | | Collided ribosome in a disome unit from S. cerevisiae | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | 登録日 | 2022-05-13 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
5DDI
 
 | |
7ZUW
 
 | | Structure of RQT (C1) bound to the stalled ribosome in a disome unit from S. cerevisiae | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | 登録日 | 2022-05-13 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
1T46
 
 | | STRUCTURAL BASIS FOR THE AUTOINHIBITION AND STI-571 INHIBITION OF C-KIT TYROSINE KINASE | | 分子名称: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Homo sapiens v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog, PHOSPHATE ION | | 著者 | Mol, C.D, Dougan, D.R, Schneider, T.R, Skene, R.J, Kraus, M.L, Scheibe, D.N, Snell, G.P, Zou, H, Sang, B.C, Wilson, K.P. | | 登録日 | 2004-04-28 | | 公開日 | 2004-06-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase.
J.Biol.Chem., 279, 2004
|
|
7ZHC
 
 | | Moss spermine/spermidine acetyl transferase (PpSSAT) in complex with AcetylCoA and polyethylen glycol | | 分子名称: | ACETYL COENZYME *A, GLYCEROL, N-acetyltransferase domain-containing protein, ... | | 著者 | Morera, S, Kopecny, D, Vigouroux, A. | | 登録日 | 2022-04-06 | | 公開日 | 2023-03-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.819 Å) | | 主引用文献 | Biochemical and structural basis of polyamine, lysine and ornithine acetylation catalyzed by spermine/spermidine N-acetyl transferase in moss and maize.
Plant J., 114, 2023
|
|
1T67
 
 | | Crystal Structure of Human HDAC8 complexed with MS-344 | | 分子名称: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, SODIUM ION, ... | | 著者 | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | 登録日 | 2004-05-05 | | 公開日 | 2004-07-27 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
4V6X
 
 | | Structure of the human 80S ribosome | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Anger, A.M, Armache, J.-P, Berninghausen, O, Habeck, M, Subklewe, M, Wilson, D.N, Beckmann, R. | | 登録日 | 2013-02-27 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Structures of the human and Drosophila 80S ribosome.
Nature, 497, 2013
|
|
6W6V
 
 | | Structure of yeast RNase MRP holoenzyme | | 分子名称: | RNA component of RNase MRP NME1, RNases MRP/P 32.9 kDa subunit, Ribonuclease MRP protein subunit RMP1, ... | | 著者 | Perederina, A, Li, D, Lee, H, Bator, C, Berezin, I, Hafenstein, S.L, Krasilnikov, A.S. | | 登録日 | 2020-03-17 | | 公開日 | 2020-07-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Cryo-EM structure of catalytic ribonucleoprotein complex RNase MRP.
Nat Commun, 11, 2020
|
|
5DHV
 
 | | HIV-1 Rev NTD dimers with variable crossing angles | | 分子名称: | Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, light chain, ... | | 著者 | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | 登録日 | 2015-08-31 | | 公開日 | 2016-06-22 | | 最終更新日 | 2017-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
6PAR
 
 | |
7AQL
 
 | |
4V58
 
 | | Crystal structure of fatty acid synthase from thermomyces lanuginosus at 3.1 angstrom resolution. | | 分子名称: | FATTY ACID SYNTHASE ALPHA SUBUNITS, FATTY ACID SYNTHASE BETA SUBUNITS, FLAVIN MONONUCLEOTIDE | | 著者 | Jenni, S, Leibundgut, M, Boehringer, D, Frick, C, Mikolasek, B, Ban, N. | | 登録日 | 2007-03-09 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structure of Fungal Fatty Acid Synthase and Implications for Iterative Substrate Shuttling
Science, 316, 2007
|
|
2VTF
 
 | | X-ray crystal structure of the Endo-beta-N-acetylglucosaminidase from Arthrobacter protophormiae E173Q mutant reveals a TIM barrel catalytic domain and two ancillary domains | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-BETA-N-ACETYLGLUCOSAMINIDASE, TRIETHYLENE GLYCOL | | 著者 | Ling, Z, Bingham, R.J, Suits, M.D.L, Moir, J.W.B, Fairbanks, A.J, Taylor, E.J. | | 登録日 | 2008-05-14 | | 公開日 | 2009-03-31 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | The X-Ray Crystal Structure of an Arthrobacter Protophormiae Endo-Beta-N-Acetylglucosaminidase Reveals a (Beta/Alpha)(8) Catalytic Domain, Two Ancillary Domains and Active Site Residues Key for Transglycosylation Activity.
J.Mol.Biol., 389, 2009
|
|
6W36
 
 | | Crystal structure of FAM46C | | 分子名称: | SULFATE ION, Terminal nucleotidyltransferase 5C | | 著者 | Shang, G.J, Zhang, X.W, Chen, H, Lu, D.F. | | 登録日 | 2020-03-09 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.854 Å) | | 主引用文献 | Structural and Functional Analyses of the FAM46C/Plk4 Complex.
Structure, 28, 2020
|
|
7AZD
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 20 bound | | 分子名称: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | 著者 | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | 登録日 | 2020-11-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
1JSZ
 
 | | Crystal Structure Analysis of N7,9-dimethylguanine-VP39 complex | | 分子名称: | 7,9-DIMETHYLGUANINE, S-ADENOSYL-L-HOMOCYSTEINE, VP39 | | 著者 | Hu, G, Oguro, A, Gershon, P.D, Quiocho, F.A. | | 登録日 | 2001-08-19 | | 公開日 | 2002-07-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | The "cap-binding slot" of an mRNA cap-binding protein: quantitative effects of aromatic side chain choice in the double-stacking sandwich with cap.
Biochemistry, 41, 2002
|
|
2W5X
 
 | | Structure of TAB5 alkaline phosphatase mutant His 135 Glu with Mg bound in the M3 site. | | 分子名称: | ALKALINE PHOSPHATASE, MAGNESIUM ION, ZINC ION | | 著者 | Koutsioulis, D, Lyskowski, A, Maki, S, Guthrie, E, Feller, G, Bouriotis, V, Heikinheimo, P. | | 登録日 | 2008-12-15 | | 公開日 | 2009-11-24 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Coordination Sphere of the Third Metal Site is Essential to the Activity and Metal Selectivity of Alkaline Phosphatases.
Protein Sci., 19, 2010
|
|
7AWK
 
 | |
1K0M
 
 | | Crystal structure of a soluble monomeric form of CLIC1 at 1.4 angstroms | | 分子名称: | CHLORIDE INTRACELLULAR CHANNEL PROTEIN 1 | | 著者 | Harrop, S.J, DeMaere, M.Z, Fairlie, W.D, Reztsova, T, Valenzuela, S.M, Mazzanti, M, Tonini, R, Qiu, M.R, Jankova, L, Warton, K, Bauskin, A.R, Wu, W.M, Pankhurst, S, Campbell, T.J, Breit, S.N, Curmi, P.M.G. | | 登録日 | 2001-09-19 | | 公開日 | 2001-12-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-A resolution.
J.Biol.Chem., 276, 2001
|
|
5D70
 
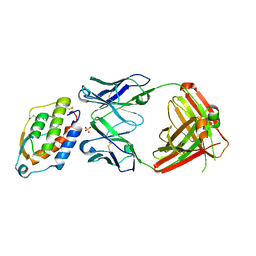 | | Crystal structure of MOR03929, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | 分子名称: | Granulocyte-macrophage colony-stimulating factor, Immunglobulin G1 Fab fragment, heavy chain, ... | | 著者 | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | 登録日 | 2015-08-13 | | 公開日 | 2015-10-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
5KCU
 
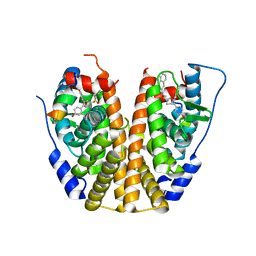 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, alpha-naphthyl OBHS-N derivative | | 分子名称: | (1S,2R,4S)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(naphthalen-2-yl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-06-07 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5D7S
 
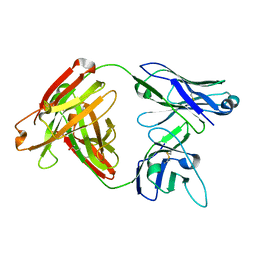 | | Crystal structure of MOR04357, a neutralizing anti-human GM-CSF antibody Fab fragment | | 分子名称: | (2S,3S)-butane-2,3-diol, Immunglobulin G1 Fab fragment, heavy chain, ... | | 著者 | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | 登録日 | 2015-08-14 | | 公開日 | 2015-10-14 | | 最終更新日 | 2016-01-13 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
7ZK9
 
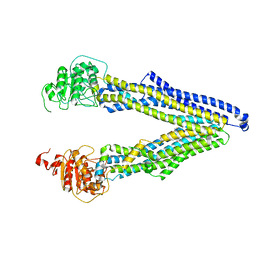 | | ABCB1 L971C mutant (mABCB1) in the inward facing state | | 分子名称: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1 | | 著者 | Parey, K, Januliene, D, Gewering, T, Zhang, Q, Moeller, A. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
2W5J
 
 | | Structure of the c14-rotor ring of the proton translocating chloroplast ATP synthase | | 分子名称: | ATP SYNTHASE C CHAIN, CHLOROPLASTIC | | 著者 | Vollmar, M, Schlieper, D, Winn, M, Buechner, C, Groth, G. | | 登録日 | 2008-12-10 | | 公開日 | 2009-05-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structure of the c14 rotor ring of the proton translocating chloroplast ATP synthase.
J. Biol. Chem., 284, 2009
|
|
5D43
 
 | |
