1AW2
 
 | | TRIOSEPHOSPHATE ISOMERASE OF VIBRIO MARINUS | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Maes, D, Zeelen, J.P, Wierenga, R.K. | | Deposit date: | 1997-10-09 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Triose-phosphate isomerase (TIM) of the psychrophilic bacterium Vibrio marinus. Kinetic and structural properties.
J.Biol.Chem., 273, 1998
|
|
5O58
 
 | | Structure of the inactive T.maritima PDE (TM1595) D80N D154N mutant with substrate 5'-pApG | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Witte, G, Drexler, D, Mueller, M. | | Deposit date: | 2017-06-01 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure, 25, 2017
|
|
1AXS
 
 | | MATURE OXY-COPE CATALYTIC ANTIBODY WITH HAPTEN | | Descriptor: | (1S,2S,5S)2-(4-GLUTARIDYLBENZYL)-5-PHENYL-1-CYCLOHEXANOL, CADMIUM ION, OXY-COPE CATALYTIC ANTIBODY | | Authors: | Mundorff, E.C, Ulrich, H.D, Stevens, R.C. | | Deposit date: | 1997-10-20 | | Release date: | 1998-02-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The interplay between binding energy and catalysis in the evolution of a catalytic antibody.
Nature, 389, 1997
|
|
3OAZ
 
 | | A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity | | Descriptor: | CHLORIDE ION, Fab 2G12, heavy chain, ... | | Authors: | Doores, K.J, Fulton, Z, Hong, V, Patel, M.K, Scanlan, C.N, Wormald, M.R, Finn, M.G, Burton, D.R, Wilson, I.A, Davis, B.G. | | Deposit date: | 2010-08-06 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5NS6
 
 | | Crystal structure of beta-glucosidase BglM-G1 from marine metagenome | | Descriptor: | Beta-glucosidase, GLYCEROL, SULFATE ION | | Authors: | Mhaindarkar, D.C, Gasper, R, Lupilova, N, Leichert, L.I, Hofmann, E. | | Deposit date: | 2017-04-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Loss of a conserved salt bridge in bacterial glycosyl hydrolase BgIM-G1 improves substrate binding in temperate environments.
Commun Biol, 1, 2018
|
|
6R47
 
 | |
6ZJO
 
 | | Crystal Structure of Staphylococcus aureus RsgA. | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Small ribosomal subunit biogenesis GTPase RsgA, ... | | Authors: | Bennison, D.J, Rafferty, J.B, Corrigan, R.M. | | Deposit date: | 2020-06-29 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Stringent Response Inhibits 70S Ribosome Formation in Staphylococcus aureus by Impeding GTPase-Ribosome Interactions.
Mbio, 12, 2021
|
|
5O60
 
 | | Structure of the 50S large ribosomal subunit from Mycobacterium smegmatis | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Hentschel, J, Burnside, C, Mignot, I, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2017-06-02 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The Complete Structure of the Mycobacterium smegmatis 70S Ribosome.
Cell Rep, 20, 2017
|
|
6ZBT
 
 | | Structure of 14-3-3 gamma in complex with Nedd4-2 14-3-3 binding motif Ser342 | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, 14-3-3 protein gamma, E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Joshi, R, Kalabova, D, Obsil, T, Obsilova, V. | | Deposit date: | 2020-06-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79948521 Å) | | Cite: | 14-3-3-protein regulates Nedd4-2 by modulating interactions between HECT and WW domains.
Commun Biol, 4, 2021
|
|
8ADU
 
 | | Lipidic alpha-synuclein fibril - polymorph L1A | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-07-11 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
5O70
 
 | |
8A9S
 
 | | Crystal structure of Ca2+-discharged obelin in complex with coelenteramine-v | | Descriptor: | 3-azanyl-2-(phenylmethyl)benzo[f]quinoxalin-8-ol, DIMETHYL SULFOXIDE, Obelin, ... | | Authors: | Kovaleva, M.I, Natashin, P.V, Schevtsov, M.B, Eremeeva, E.V, Bukhdruker, S.S, Dmitrieva, D.A, Mishin, A.V, Vysotski, E.S, Borshchevskiy, V.I. | | Deposit date: | 2022-06-29 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of semi-synthetic obelin-v after calcium induced bioluminescence implies coelenteramine as the main reaction product.
Sci Rep, 12, 2022
|
|
6TMY
 
 | | Crystal structure of isoform CBd of the basic phospholipase A2 subunit of crotoxin from Crotalus durissus terrificus | | Descriptor: | CHLORIDE ION, Phospholipase A2 crotoxin basic subunit CBc, SODIUM ION, ... | | Authors: | Nemecz, D, Ostrowski, M, Saul, F.A, Faure, G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Isoform CBd of the Basic Phospholipase A 2 Subunit of Crotoxin: Description of the Structural Framework of CB for Interaction with Protein Targets.
Molecules, 25, 2020
|
|
4UIK
 
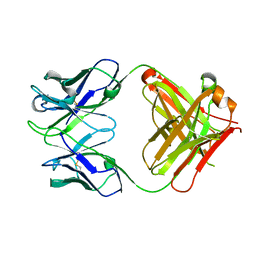 | | crystal structure of quinine-dependent Fab 314.1 | | Descriptor: | FAB 314.1 | | Authors: | Zhu, J, Zhu, J, Bougie, D.W, Aster, R.H, Springer, T.A. | | Deposit date: | 2015-03-30 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Quinine-Dependent Antibody Binding to Platelet Integrin Alphaiib Beta3
Blood, 126, 2015
|
|
6Z01
 
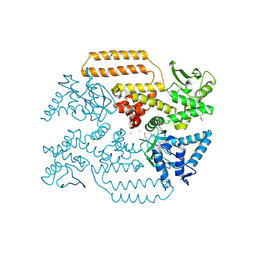 | | DNA Topoisomerase | | Descriptor: | CHLORIDE ION, DNA topoisomerase I | | Authors: | Takahashi, T.S, Gadelle, D, Forterre, P, Mayer, C, Petrella, S. | | Deposit date: | 2020-05-07 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Topoisomerase I (TOP1) dynamics: conformational transition from open to closed states.
Nat Commun, 13, 2022
|
|
6Z03
 
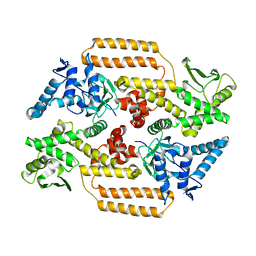 | | DNA Topoisomerase | | Descriptor: | DNA topoisomerase I | | Authors: | Takahashi, T.S, Gadelle, D, Forterre, P, Mayer, C, Petrella, S. | | Deposit date: | 2020-05-07 | | Release date: | 2021-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Topoisomerase I (TOP1) dynamics: conformational transition from open to closed states.
Nat Commun, 13, 2022
|
|
3HS7
 
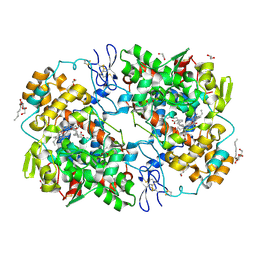 | | X-ray crystal structure of docosahexaenoic acid bound to the cyclooxygenase channel of cyclooxygenase-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Simmons, D.M, Malkowski, M.G. | | Deposit date: | 2009-06-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of fatty acid substrate binding to cyclooxygenase-2.
J.Biol.Chem., 285, 2010
|
|
6Z3E
 
 | |
7RY9
 
 | | S. CEREVISIAE CYP51 I471T mutant COMPLEXED WITH Voriconazole | | Descriptor: | Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE, TETRAETHYLENE GLYCOL, ... | | Authors: | Graham, D.O, Wilson, R.K, Ruma, Y.N, Keniya, M.V, Tyndall, J.D, Monk, B.C. | | Deposit date: | 2021-08-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Azole Resistance of the Candida albicans Darlington Strain Using Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase as a Surrogate.
J Fungi (Basel), 7, 2021
|
|
5O9O
 
 | | Crystal structure of ScGas2 in complex with compound 7. | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-[4-(naphthalen-2-ylmethoxymethyl)-1,2,3-triazol-1-yl]-3,5-bis(oxidanyl)oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2 | | Authors: | Delso, I, Valero-Gonzalez, J, Fang, W, Gomollon-Bel, F, Castro-Lopez, J, Navratilova, I, van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
1B8A
 
 | | ASPARTYL-TRNA SYNTHETASE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PROTEIN (ASPARTYL-TRNA SYNTHETASE) | | Authors: | Schmitt, E, Moulinier, L, Thierry, J.-C, Moras, D. | | Deposit date: | 1999-01-27 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation.
EMBO J., 17, 1998
|
|
6TQ7
 
 | | Crystal structure of the Orexin-1 receptor in complex with SB-334867 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 1-(2-methyl-1,3-benzoxazol-6-yl)-3-(1,5-naphthyridin-4-yl)urea, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6636 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
7RYB
 
 | | S. CEREVISIAE CYP51 Y140H/I471T - double mutant COMPLEXED WITH Voriconazole | | Descriptor: | Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE, Voriconazole | | Authors: | Graham, D.O, Wilson, R.K, Ruma, Y.N, Keniya, M.V, Tyndall, J.D, Monk, B.C. | | Deposit date: | 2021-08-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights into the Azole Resistance of the Candida albicans Darlington Strain Using Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase as a Surrogate.
J Fungi (Basel), 7, 2021
|
|
8ADS
 
 | | Lipidic alpha-synuclein fibril - polymorph L2B | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-07-11 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
6RAH
 
 | | Heterodimeric ABC exporter TmrAB in ATP-bound outward-facing open conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug resistance ABC transporter ATP-binding and permease protein, ... | | Authors: | Thomas, C, Januliene, D, Mehdipour, A.R, Hofmann, S, Hummer, G, Moeller, A, Tampe, R. | | Deposit date: | 2019-04-06 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conformation space of a heterodimeric ABC exporter under turnover conditions.
Nature, 571, 2019
|
|
