6BP4
 
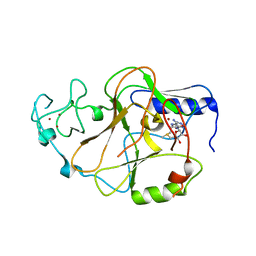 | | Structure of the S. pombe Clr4 catalytic domain bound to SAM | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Currie, M.A, Moazed, D. | | Deposit date: | 2017-11-21 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7701 Å) | | Cite: | Automethylation-induced conformational switch in Clr4 (Suv39h) maintains epigenetic stability.
Nature, 560, 2018
|
|
6BOX
 
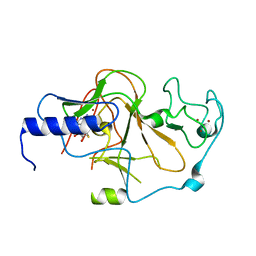 | | Structure of the S. pombe Clr4 catalytic domain bound to SAH | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Currie, M.A, Moazed, D. | | Deposit date: | 2017-11-21 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Automethylation-induced conformational switch in Clr4 (Suv39h) maintains epigenetic stability.
Nature, 560, 2018
|
|
4FL4
 
 | | Scaffoldin conformation and dynamics revealed by a ternary complex from the Clostridium thermocellum cellulosome | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Glycoside hydrolase family 9, ... | | Authors: | Currie, M.A, Adams, J.J, Faucher, F, Bayer, E.A, Jia, Z, Smith, S.P, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Scaffoldin Conformation and Dynamics Revealed by a Ternary Complex from the Clostridium thermocellum Cellulosome.
J.Biol.Chem., 287, 2012
|
|
7MFS
 
 | |
7MQT
 
 | |
7MFN
 
 | |
7T3E
 
 | | Structure of the sialic acid bound Tripartite ATP-independent Periplasmic (TRAP) periplasmic component SiaP from Photobacterium profundum | | Descriptor: | N-acetyl-beta-neuraminic acid, SULFATE ION, TRAP-type C4-dicarboxylate transport system, ... | | Authors: | Davies, J.S, Currie, M.J, North, R.A, Dobson, R.C.J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter.
Nat Commun, 14, 2023
|
|
8THJ
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (antiparallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
8THI
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (parallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, SODIUM ION, Sialic acid TRAP transporter permease protein SiaT | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
3KCP
 
 | | Crystal structure of interacting Clostridium thermocellum multimodular components | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cellulosomal-scaffolding protein A, ... | | Authors: | Adams, J.J, Currie, M.A, Bayer, E.A, Jia, Z, Smith, S.P. | | Deposit date: | 2009-10-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Insights into Higher-Order Organization of the Cellulosome Revealed by a Dissect-and-Build Approach: Crystal Structure of Interacting Clostridium thermocellum Multimodular Components
J.Mol.Biol., 396, 2010
|
|
4JJN
 
 | | Crystal structure of heterochromatin protein Sir3 in complex with a silenced yeast nucleosome | | Descriptor: | DNA (146-MER), Histone H2A.2, Histone H2B.2, ... | | Authors: | Wang, F, Li, G, Mohammed, A, Lu, C, Currie, M, Johnson, A, Moazed, D. | | Deposit date: | 2013-03-08 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8UUB
 
 | | Structure of hypothiocyanous acid reductase (Har) from Streptococcus pneumoniae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, Oxidoreductase, ... | | Authors: | Shearer, H.L, Currie, M.J, Dickerhof, N, Dobson, R.C.J. | | Deposit date: | 2023-10-31 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hypothiocyanous acid reductase is critical for host colonization and infection by Streptococcus pneumoniae.
J.Biol.Chem., 300, 2024
|
|
6VVA
 
 | |
4AQC
 
 | | Triazolopyridine-based Inhibitor of Janus Kinase 2 | | Descriptor: | 8-(4-methylsulfonylphenyl)-N-(4-morpholin-4-ylphenyl)-[1,2,4]triazolo[1,5-a]pyridin-2-amine, SULFATE ION, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Dugan, B.J, Gingrich, D.E, Mesaros, E.F, Milkiewicz, K.L, Curry, M.A, Zulli, A.L, Dobrzanski, P, Serdikoff, C, Jan, M, Angeles, T.S, Albom, M.S, Mason, J.L, Aimone, L.D, Meyer, S.L, Huang, Z, Wells-Knecht, K.J, Ator, M.A, Ruggeri, B.A, Dorsey, B.D. | | Deposit date: | 2012-04-16 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Selective, Orally Bioavailable 1,2,4-Triazolo[1,5-A]Pyridine-Based Inhibitor of Janus Kinase 2 for Use in Anticancer Therapy: Discovery of Cep-33779.
J.Med.Chem., 55, 2012
|
|
