2WDS
 
 | | Crystal structure of the Streptomyces coelicolor H110A AcpS mutant in complex with cofactor CoA at 1.3 A | | Descriptor: | COENZYME A, HOLO-[ACYL-CARRIER-PROTEIN] SYNTHASE, MAGNESIUM ION | | Authors: | Dall'Aglio, P, Arthur, C, Crump, M.P, Crosby, J, Hadfield, A.T. | | Deposit date: | 2009-03-26 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Analysis of Streptomyces Coelicolor Phosphopantetheinyl Transferase, Acps, Reveals the Basis for Relaxed Substrate Specificity.
Biochemistry, 50, 2011
|
|
2WDO
 
 | | Crystal structure of the S. coelicolor AcpS in complex with acetyl- CoA at 1.5 A | | Descriptor: | ACETYL COENZYME *A, COENZYME A, GLYCEROL, ... | | Authors: | Dall'Aglio, P, Arthur, C, Crump, M.P, Crosby, J, Hadfield, A.T. | | Deposit date: | 2009-03-25 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Analysis of Streptomyces Coelicolor Phosphopantetheinyl Transferase, Acps, Reveals the Basis for Relaxed Substrate Specificity.
Biochemistry, 50, 2011
|
|
2WDY
 
 | | Crystal structure of the Streptomyces coelicolor D111A AcpS mutant in complex with cofactor CoA at 1.4 A | | Descriptor: | COENZYME A, HOLO-[ACYL-CARRIER-PROTEIN] SYNTHASE, MAGNESIUM ION, ... | | Authors: | Dall'Aglio, P, Arthur, C, Crump, M.P, Crosby, J, Hadfield, A.T. | | Deposit date: | 2009-03-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Analysis of Streptomyces Coelicolor Phosphopantetheinyl Transferase, Acps, Reveals the Basis for Relaxed Substrate Specificity.
Biochemistry, 50, 2011
|
|
4XSA
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4XS7
 
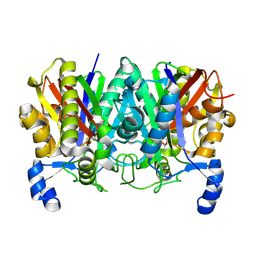 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4XS9
 
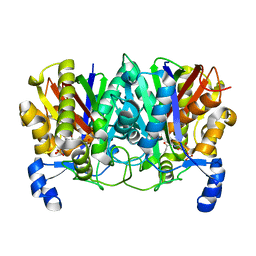 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase, N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-[2-(propanoylamino)ethyl]-beta-alaninamide | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4XSB
 
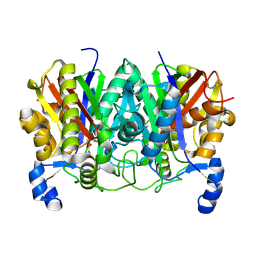 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
5LO4
 
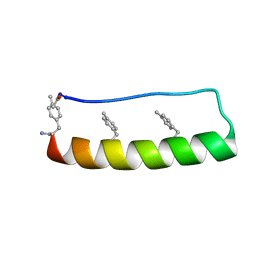 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPa-CH3 | | Authors: | Baker, E.G, Williams, C, Hudson, K.L, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
1NQ4
 
 | | Solution Structure of Oxytetracycline Acyl Carrier Protein | | Descriptor: | Oxytetracycline polyketide synthase acyl carrier protein | | Authors: | Findlow, S.C, Winsor, C, Simpson, T.J, Crosby, J, Crump, M.P. | | Deposit date: | 2003-01-21 | | Release date: | 2003-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of oxytetracycline polyketide synthase acyl carrier protein from Streptomyces rimosus
Biochemistry, 42, 2003
|
|
6Z30
 
 | | Human cation-independent mannose 6-phosphate/ IGF2 receptor domains 9-10 | | Descriptor: | Cation-independent mannose-6-phosphate receptor, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bochel, A.J, Williams, C, Crump, M.P. | | Deposit date: | 2020-05-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7-11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9.
Structure, 28, 2020
|
|
6Z31
 
 | |
6Z32
 
 | | Human cation-independent mannose 6-phosphate/IGF2 receptor domains 7-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, SULFATE ION, ... | | Authors: | Bochel, A.J, Williams, C, McCoy, A.J, Hoppe, H, Winter, A.J, Nicholls, R.D, Harlos, K, Jones, Y.E, Berger, I, Hassan, B, Crump, M.P. | | Deposit date: | 2020-05-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7-11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9.
Structure, 28, 2020
|
|
1A93
 
 | | NMR SOLUTION STRUCTURE OF THE C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MAX PROTEIN, MYC PROTO-ONCOGENE PROTEIN | | Authors: | Lavigne, P, Crump, M.P, Gagne, S.M, Hodges, R.S, Kay, C.M, Sykes, B.D. | | Deposit date: | 1998-04-15 | | Release date: | 1998-10-21 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper.
J.Mol.Biol., 281, 1998
|
|
1W4Z
 
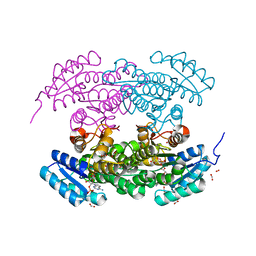 | | Structure of actinorhodin polyketide (actIII) Reductase | | Descriptor: | FORMIC ACID, KETOACYL REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hadfield, A.T, Limpkin, C, Teartasin, W, Simpson, T.J, Crosby, J, Crump, M.P. | | Deposit date: | 2004-08-03 | | Release date: | 2004-12-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of the Actiii Actinorhodin Polyketide Reductase; Proposed Mechanism for Acp and Polyketide Binding
Structure, 12, 2004
|
|
6SZC
 
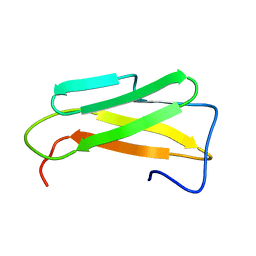 | |
2A93
 
 | | NMR SOLUTION STRUCTURE OF THE C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER, 40 STRUCTURES | | Descriptor: | C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER | | Authors: | Lavigne, P, Crump, M.P, Gagne, S.M, Hodges, R.S, Kay, C.M, Sykes, B.D. | | Deposit date: | 1998-06-09 | | Release date: | 1999-01-27 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper.
J.Mol.Biol., 281, 1998
|
|
4C26
 
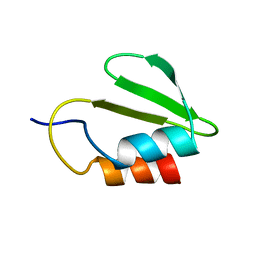 | | Solution NMR structure of the HicA toxin from Burkholderia pseudomallei | | Descriptor: | HICA | | Authors: | Butt, A, Higman, V.A, Williams, C, Crump, M.P, Hemsley, C, Harmer, N, Titball, R.W. | | Deposit date: | 2013-08-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Hica Toxin from Burkholderia Pseudomallei Has a Role in Persister Cell Formation.
Biochem.J., 459, 2014
|
|
2CNR
 
 | |
5LO2
 
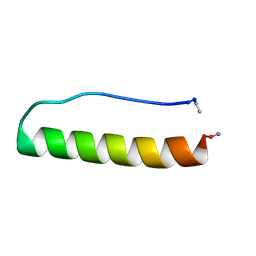 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPaTyr | | Authors: | Baker, E.G, Hudson, K.L, Williams, C, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
5LO3
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPaOMe | | Authors: | Baker, E.G, Hudson, K.L, Williams, C, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
5NOC
 
 | |
6G1C
 
 | |
6FXD
 
 | | Crystal structure of MupZ from Pseudomonas fluorescens | | Descriptor: | MupZ | | Authors: | Wang, L, Parnell, A, Williams, C, Bakar, N.A, van der Kamp, M.W, Simpson, T.J, Race, P.R, Crump, M.P, Willis, C.L. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Rieske oxygenase/epoxide hydrolase-catalysed reaction cascade creates oxygen heterocycles in mupirocin biosynthesis
Nat Catal, 2018
|
|
6G1N
 
 | |
6GWX
 
 | | Stabilising and Understanding a Miniprotein by Rational Design. | | Descriptor: | Optimised PPa-TYR | | Authors: | Porter Goff, K.L, Williams, C, Baker, E.G, Nicol, D, Samphire, J.L, Zieleniewski, F.L, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2018-06-26 | | Release date: | 2019-07-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Stabilizing and Understanding a Miniprotein by Rational Redesign.
Biochemistry, 58, 2019
|
|
