6TG5
 
 | |
1A93
 
 | | NMR SOLUTION STRUCTURE OF THE C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | MAX PROTEIN, MYC PROTO-ONCOGENE PROTEIN | | 著者 | Lavigne, P, Crump, M.P, Gagne, S.M, Hodges, R.S, Kay, C.M, Sykes, B.D. | | 登録日 | 1998-04-15 | | 公開日 | 1998-10-21 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper.
J.Mol.Biol., 281, 1998
|
|
6G26
 
 | | The crystal structure of the Burkholderia pseudomallei HicAB complex | | 分子名称: | 1,2-ETHANEDIOL, HicA, HicB, ... | | 著者 | Winter, A.J, Isupov, M.N, Williams, C, Crump, M.P. | | 登録日 | 2018-03-22 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | The molecular basis of protein toxin HicA-dependent binding of the protein antitoxin HicB to DNA.
J. Biol. Chem., 293, 2018
|
|
5NOC
 
 | |
6G1N
 
 | |
7OUB
 
 | |
6G1C
 
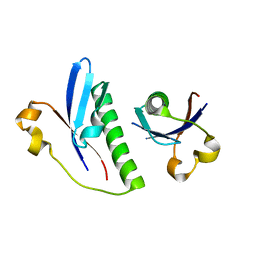 | |
2A93
 
 | | NMR SOLUTION STRUCTURE OF THE C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER, 40 STRUCTURES | | 分子名称: | C-MYC-MAX HETERODIMERIC LEUCINE ZIPPER | | 著者 | Lavigne, P, Crump, M.P, Gagne, S.M, Hodges, R.S, Kay, C.M, Sykes, B.D. | | 登録日 | 1998-06-09 | | 公開日 | 1999-01-27 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Insights into the mechanism of heterodimerization from the 1H-NMR solution structure of the c-Myc-Max heterodimeric leucine zipper.
J.Mol.Biol., 281, 1998
|
|
6Z31
 
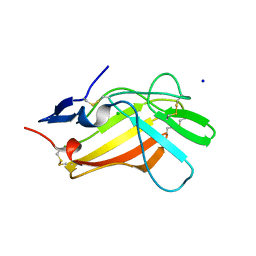 | |
6Z32
 
 | | Human cation-independent mannose 6-phosphate/IGF2 receptor domains 7-11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-independent mannose-6-phosphate receptor, SULFATE ION, ... | | 著者 | Bochel, A.J, Williams, C, McCoy, A.J, Hoppe, H, Winter, A.J, Nicholls, R.D, Harlos, K, Jones, Y.E, Berger, I, Hassan, B, Crump, M.P. | | 登録日 | 2020-05-19 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.47 Å) | | 主引用文献 | Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7-11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9.
Structure, 28, 2020
|
|
6Z30
 
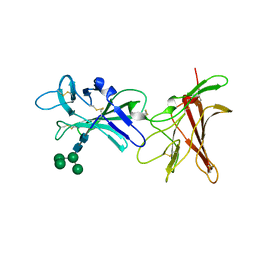 | | Human cation-independent mannose 6-phosphate/ IGF2 receptor domains 9-10 | | 分子名称: | Cation-independent mannose-6-phosphate receptor, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Bochel, A.J, Williams, C, Crump, M.P. | | 登録日 | 2020-05-19 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7-11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9.
Structure, 28, 2020
|
|
6FXD
 
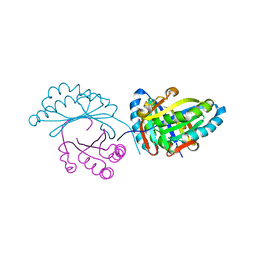 | | Crystal structure of MupZ from Pseudomonas fluorescens | | 分子名称: | MupZ | | 著者 | Wang, L, Parnell, A, Williams, C, Bakar, N.A, van der Kamp, M.W, Simpson, T.J, Race, P.R, Crump, M.P, Willis, C.L. | | 登録日 | 2018-03-08 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | A Rieske oxygenase/epoxide hydrolase-catalysed reaction cascade creates oxygen heterocycles in mupirocin biosynthesis
Nat Catal, 2018
|
|
4C26
 
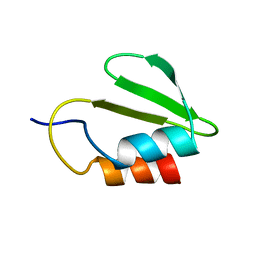 | | Solution NMR structure of the HicA toxin from Burkholderia pseudomallei | | 分子名称: | HICA | | 著者 | Butt, A, Higman, V.A, Williams, C, Crump, M.P, Hemsley, C, Harmer, N, Titball, R.W. | | 登録日 | 2013-08-16 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Hica Toxin from Burkholderia Pseudomallei Has a Role in Persister Cell Formation.
Biochem.J., 459, 2014
|
|
5TT4
 
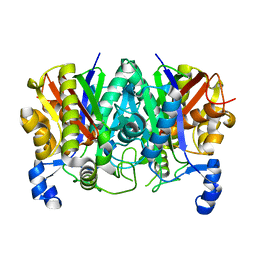 | | Determining the Molecular Basis For Starter Unit Selection During Daunorubicin Biosynthesis | | 分子名称: | Daunorubicin-doxorubicin polyketide synthase | | 著者 | Jackson, D.R, Valentic, T.R, Patel, A, Tsai, S.C, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | 登録日 | 2016-11-01 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Determining the Molecular Basis For Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4XS7
 
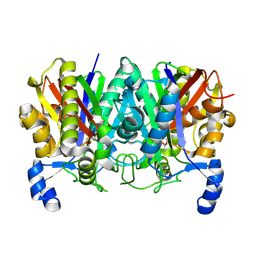 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | 分子名称: | Daunorubicin-doxorubicin polyketide synthase | | 著者 | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | 登録日 | 2015-01-22 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4XSA
 
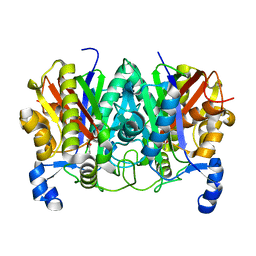 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | 分子名称: | Daunorubicin-doxorubicin polyketide synthase | | 著者 | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | 登録日 | 2015-01-22 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.204 Å) | | 主引用文献 | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4XSB
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | 分子名称: | Daunorubicin-doxorubicin polyketide synthase | | 著者 | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | 登録日 | 2015-01-22 | | 公開日 | 2016-01-27 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.203 Å) | | 主引用文献 | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4XS9
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | 分子名称: | Daunorubicin-doxorubicin polyketide synthase, N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-[2-(propanoylamino)ethyl]-beta-alaninamide | | 著者 | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | 登録日 | 2015-01-22 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
6SZC
 
 | |
2CNJ
 
 | | NMR studies on the interaction of Insulin-Growth Factor II (IGF-II) with IGF2R domain 11 | | 分子名称: | CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | 著者 | Williams, C, Prince, S, Zaccheo, O, Hassan, A.B, Crosby, J, Crump, M. | | 登録日 | 2006-05-22 | | 公開日 | 2007-05-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Insights Into the Interaction of Insulin-Like Growth Factor 2 with Igf2R Domain 11.
Structure, 15, 2007
|
|
2LLA
 
 | | NMR solution structure ensemble of domain 11 of the echidna M6P/IGF2R receptor | | 分子名称: | Mannose-6-phosphate/insulin-like growth factor II receptor | | 著者 | Strickland, M, Crump, M.P, Williams, C, Rezgui, D, Ellis, R.Z, Hoppe, H, Frago, S, Prince, S.N, Zaccheo, O.J, Forbes, B.E, Jones, E, Hassan, A.Z, Wattana-Amorn, P. | | 登録日 | 2011-11-05 | | 公開日 | 2012-11-07 | | 最終更新日 | 2012-12-12 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An exon splice enhancer primes IGF2:IGF2R binding site structure and function evolution.
Science, 338, 2012
|
|
5LO4
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | 分子名称: | PPa-CH3 | | 著者 | Baker, E.G, Williams, C, Hudson, K.L, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | 登録日 | 2016-08-08 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
5LO2
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | 分子名称: | PPaTyr | | 著者 | Baker, E.G, Hudson, K.L, Williams, C, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | 登録日 | 2016-08-08 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
2M6T
 
 | | NMR solution structure ensemble of 3-4D mutant domain 11 IGF2R | | 分子名称: | Insulin-like growth factor 2 receptor variant | | 著者 | Strickland, M, Williams, C, Richards, E, Minnall, L, Crump, M.P, Frago, S, Hughes, J, Garner, L, Hoppe, H, Rezgui, D, Zaccheo, O.J, Prince, S.N, Hassan, A.B, Whittaker, S. | | 登録日 | 2013-04-09 | | 公開日 | 2014-10-15 | | 最終更新日 | 2016-06-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Functional evolution of IGF2:IGF2R domain 11 binding generates novel structural interactions and a specific IGF2 antagonist.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5LO3
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | 分子名称: | PPaOMe | | 著者 | Baker, E.G, Hudson, K.L, Williams, C, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | 登録日 | 2016-08-08 | | 公開日 | 2017-05-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
