1H0B
 
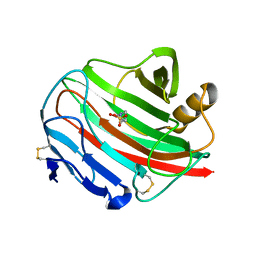 | | Endoglucanase cel12A from Rhodothermus marinus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CELLULASE | | Authors: | Crennell, S.J, Hreggvidsson, G.O, Nordberg-Karlsson, E. | | Deposit date: | 2002-06-17 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Rhodothermus Marinus Cel12A, a Highly Thermostable Family 12 Endoglucanase, at 1.8 A Resolution
J.Mol.Biol., 320, 2002
|
|
5J78
 
 | | Crystal structure of an Acetylating Aldehyde Dehydrogenase from Geobacillus thermoglucosidasius | | Descriptor: | ACETATE ION, Acetaldehyde dehydrogenase (Acetylating), GLYCEROL, ... | | Authors: | Crennell, S.J, Extance, J.P, Danson, M.J. | | Deposit date: | 2016-04-06 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an acetylating aldehyde dehydrogenase from the thermophilic ethanologen Geobacillus thermoglucosidasius.
Protein Sci., 25, 2016
|
|
5J7I
 
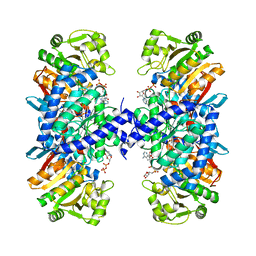 | |
2YDA
 
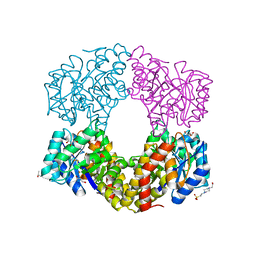 | | Sulfolobus sulfataricus 2-keto-3-deoxygluconate aldolase Y103F,Y130F, A198F variant | | Descriptor: | 2-KETO-3-DEOXY GLUCONATE ALDOLASE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Crennell, S.J, Royer, S.F, Angelopoulou, M, Hough, D.W, Danson, M.J, Bull, S.D. | | Deposit date: | 2011-03-18 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Sulfolobus Sulfataricus 2-Keto-3-Deoxygluconate Aldolase
To be Published
|
|
2BWA
 
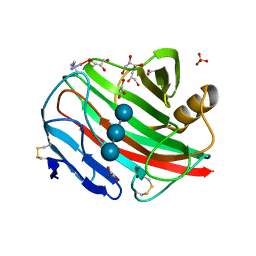 | |
2BWC
 
 | |
2BW8
 
 | |
6FB4
 
 | | human KIBRA C2 domain mutant C771A | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein KIBRA | | Authors: | Crennell, S.J, Posner, M.G, Bagby, S. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.415631 Å) | | Cite: | Distinctive phosphoinositide- and Ca2+-binding properties of normal and cognitive performance-linked variant forms of KIBRA C2 domain.
J. Biol. Chem., 293, 2018
|
|
6FJD
 
 | | Human KIBRA C2 domain mutant C771A in complex with phosphatidylinositol 4,5-bisphosphate | | Descriptor: | (2R)-3-{[(R)-HYDROXY{[(1R,2R,3S,4R,5R,6S)-2,3,6-TRIHYDROXY-4,5-BIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL]OXY}PROPANE-1 ,2-DIYL DIBUTANOATE, GLYCEROL, Protein KIBRA, ... | | Authors: | Crennell, S.J, Posner, M.G, Bagby, S. | | Deposit date: | 2018-01-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Distinctive phosphoinositide- and Ca2+-binding properties of normal and cognitive performance-linked variant forms of KIBRA C2 domain.
J. Biol. Chem., 293, 2018
|
|
6FJC
 
 | | Human KIBRA C2 domain mutant C771A in complex with phosphatidylinositol 3,4,5-trisphosphate | | Descriptor: | (2R)-3-{[(S)-{[(2S,3R,5S,6S)-2,6-DIHYDROXY-3,4,5-TRIS(PHOSPHONOOXY)CYCLOHEXYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-(1-HYDROXY BUTOXY)PROPYL BUTYRATE, GLYCEROL, Protein KIBRA, ... | | Authors: | Crennell, S.J, Posner, M.G, Bagby, S. | | Deposit date: | 2018-01-22 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Distinctive phosphoinositide- and Ca2+-binding properties of normal and cognitive performance-linked variant forms of KIBRA C2 domain.
J. Biol. Chem., 293, 2018
|
|
6FD0
 
 | | Human KIBRA C2 domain mutant M734I S735A | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Protein KIBRA | | Authors: | Crennell, S.J, Posner, M.G, Bagby, S. | | Deposit date: | 2017-12-21 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.64215851 Å) | | Cite: | Distinctive phosphoinositide- and Ca2+-binding properties of normal and cognitive performance-linked variant forms of KIBRA C2 domain.
J. Biol. Chem., 293, 2018
|
|
6G3Z
 
 | | Sulfolobus sulfataricus 2-keto-3-deoxygluconate (KDG) aldolase complex with D-KDPG | | Descriptor: | 2-dehydro-3-deoxy-phosphogluconate/2-dehydro-3-deoxy-6-phosphogalactonate aldolase, 2-keto 3 deoxy 6 phospho gluconate, ISOPROPYL ALCOHOL | | Authors: | Crennell, S.J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into the Substrate Specificity of Archaeal Entner-Doudoroff Aldolases: The Structures of Picrophilus torridus 2-Keto-3-deoxygluconate Aldolase and Sulfolobus solfataricus 2-Keto-3-deoxy-6-phosphogluconate Aldolase in Complex with 2-Keto-3-deoxy-6-phosphogluconate.
Biochemistry, 57, 2018
|
|
6GT8
 
 | |
6H2S
 
 | |
6GV2
 
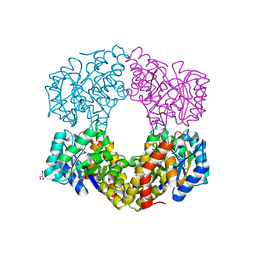 | | Sulfolobus solfataricus 2-keto-3-deoxygluconate aldolase Y103F,Y130F,A198F variant in complex with L-2-keto, 3-deoxy-galactonate | | Descriptor: | 1,2-ETHANEDIOL, 2-dehydro-3-deoxy-phosphogluconate/2-dehydro-3-deoxy-6-phosphogalactonate aldolase, 3-DEOXY-D-ARABINO-HEXONIC ACID, ... | | Authors: | Crennell, S.J, Danson, M.J, Royer, S. | | Deposit date: | 2018-06-20 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Sulfolobus solfataricus 2-keto-3-deoxygluconate aldolase Y132V,T157C variant
To Be Published
|
|
6GSO
 
 | | Sulfolobus solfataricus 2-keto-3-deoxygluconate aldolase Y132V,T157C variant | | Descriptor: | 1,2-ETHANEDIOL, 2-dehydro-3-deoxy-phosphogluconate/2-dehydro-3-deoxy-6-phosphogalactonate aldolase, GLYCEROL | | Authors: | Crennell, S.J, Danson, M.J, Royer, S. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfolobus solfataricus 2-keto-3-deoxygluconate aldolase Y132V,T157C variant
To Be Published
|
|
6H7S
 
 | |
6H2R
 
 | |
6H7R
 
 | |
4UXD
 
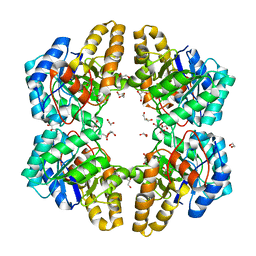 | | 2-keto 3-deoxygluconate aldolase from Picrophilus torridus | | Descriptor: | 1,2-ETHANEDIOL, 2-DEHYDRO-3-DEOXY-D-GLUCONATE/2-DEHYDRO-3-DEOXY-PHOSPHOGLUCONATE ALDOLASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Priftis, A, Zaitsev, V, Reher, M, Johnsen, U, Danson, M.J, Taylor, G.L, Schoenheit, P, Crennell, S.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the Substrate Specificity of Archaeal Entner-Doudoroff Aldolases: The Structures of Picrophilus torridus 2-Keto-3-deoxygluconate Aldolase and Sulfolobus solfataricus 2-Keto-3-deoxy-6-phosphogluconate Aldolase in Complex with 2-Keto-3-deoxy-6-phosphogluconate.
Biochemistry, 57, 2018
|
|
3ZDR
 
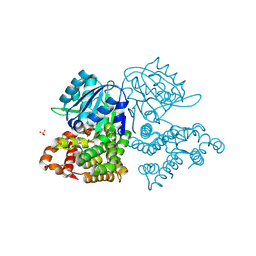 | | Structure of the Alcohol dehydrogenase (ADH) domain of a bifunctional ADHE dehydrogenase from Geobacillus thermoglucosidasius NCIMB 11955 | | Descriptor: | ALCOHOL DEHYDROGENASE DOMAIN OF THE BIFUNCTIONAL ACETALDEHYDE DEHYDROGENASE, GLYCEROL, SULFATE ION, ... | | Authors: | Extance, J, Crennell, S.J, Eley, K, Cripps, R, Hough, D.W, Danson, M.J. | | Deposit date: | 2012-11-30 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structure of a Bifunctional Alcohol Dehydrogenase Involved in Bioethanol Generation in Geobacillus Thermoglucosidasius
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6RMT
 
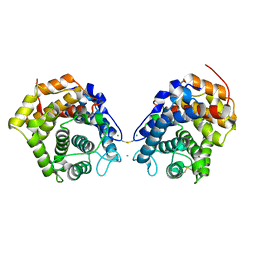 | |
6RMU
 
 | | Crystal structure of disulphide-linked human C3d dimer in complex with Staphylococcus aureus complement subversion protein Sbi-IV | | Descriptor: | 1,2-ETHANEDIOL, Complement C3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wahid, A.A, van den Elsen, J.M.H, Crennell, S.J. | | Deposit date: | 2019-05-07 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Staphylococcal Complement Evasion Protein Sbi Stabilises C3d Dimers by Inducing an N-Terminal Helix Swap
Front Immunol, 13, 2022
|
|
7Z5Q
 
 | |
4OFS
 
 | | Crystal structure of a truncated catalytic core of the 2-oxoacid dehydrogenase multienzyme complex from Thermoplasma acidophilum | | Descriptor: | Probable lipoamide acyltransferase | | Authors: | Marrot, N.L, Marshall, J.J.T, Svergun, D.I, Crennell, S.J, Hough, D.W, van den Elsen, J.M.H, Danson, M.J. | | Deposit date: | 2014-01-15 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Why are the 2-oxoacid dehydrogenase complexes so large? Generation of an active trimeric complex.
Biochem.J., 463, 2014
|
|
