7ZSB
 
 | |
7ZSA
 
 | |
7ZS9
 
 | |
7ZWC
 
 | | Structure of SNAPc:TBP-TFIIA-TFIIB sub-complex bound to U5 snRNA promoter | | Descriptor: | Non-template strand, TATA-box-binding protein, Template strand, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1Q1H
 
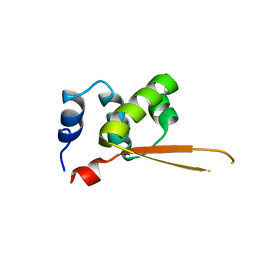 | |
8BVW
 
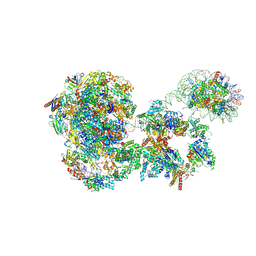 | | RNA polymerase II pre-initiation complex with the distal +1 nucleosome (PIC-Nuc18W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-20 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
8B3I
 
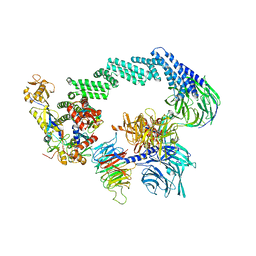 | | CRL4CSA-E2-Ub (state 2) | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | C(N)RL4CSA-E2-Ub (state 2)
To Be Published
|
|
8B3G
 
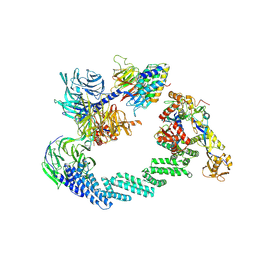 | | C(N)RL4CSA-UVSSA-E2-ubiquitin complex. | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2022-09-16 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for RNA polymerase II ubiquitylation and inactivation in transcription-coupled repair.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8CEO
 
 | |
8CEN
 
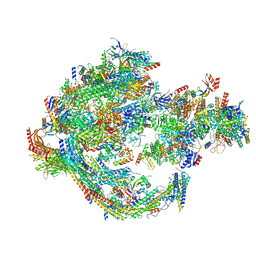 | | Yeast RNA polymerase II transcription pre-initiation complex with core Mediator | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Wang, H, Schilbach, S, Cramer, P. | | Deposit date: | 2023-02-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Yeast PIC-Mediator structure with RNA polymerase II C-terminal domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8BYQ
 
 | | RNA polymerase II pre-initiation complex with the proximal +1 nucleosome (PIC-Nuc10W) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
5L3X
 
 | |
8BZ1
 
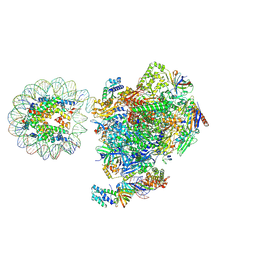 | | RNA polymerase II core pre-initiation complex with the proximal +1 nucleosome (cPIC-Nuc10W) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Abril-Garrido, J, Dienemann, C, Grabbe, F, Velychko, T, Lidschreiber, M, Wang, H, Cramer, P. | | Deposit date: | 2022-12-14 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of transcription reduction by a promoter-proximal +1 nucleosome.
Mol.Cell, 83, 2023
|
|
2XNF
 
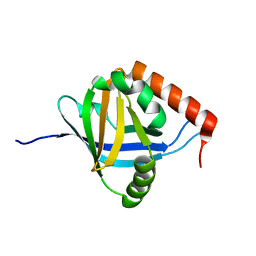 | | The Mediator Med25 activator interaction domain: Structure and cooperative binding of VP16 subdomains | | Descriptor: | MEDIATOR OF RNA POLYMERASE II TRANSCRIPTION SUBUNIT 25 | | Authors: | Vojnic, E, Mourao, A, Seizl, M, Simon, B, Wenzeck, L, Lariviere, L, Baumli, S, Meisterernst, M, Sattler, M, Cramer, P. | | Deposit date: | 2010-08-02 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Vp16 Binding of the Mediator Med25 Activator Interaction Domain.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3FBI
 
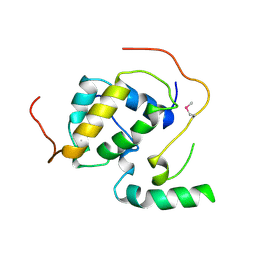 | | Structure of the Mediator submodule Med7N/31 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 31, Mediator of RNA polymerase II transcription subunit 7 | | Authors: | Koschubs, T, Seizl, M, Lariviere, L, Kurth, F, Baumli, S, Martin, D.E, Cramer, P. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification, structure, and functional requirement of the Mediator submodule Med7N/31
Embo J., 28, 2009
|
|
3FBN
 
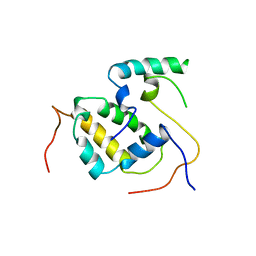 | | Structure of the Mediator submodule Med7N/31 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 31, Mediator of RNA polymerase II transcription subunit 7 | | Authors: | Koschubs, T, Seizl, M, Lariviere, L, Kurth, F, Baumli, S, Martin, D.E, Cramer, P. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.007 Å) | | Cite: | Identification, structure, and functional requirement of the Mediator submodule Med7N/31
Embo J., 28, 2009
|
|
3LPE
 
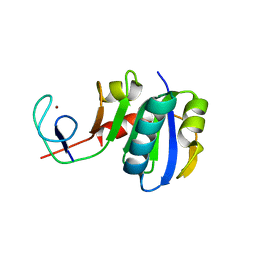 | | Crystal structure of Spt4/5NGN heterodimer complex from Methanococcus jannaschii | | Descriptor: | DNA-directed RNA polymerase subunit E'', Putative transcription antitermination protein nusG, ZINC ION | | Authors: | Hirtreiter, A, Damsma, G.E, Cheung, A.C.M, Klose, D, Grohmann, D, Vojnic, E, Martin, A.C.R, Cramer, P, Werner, F. | | Deposit date: | 2010-02-05 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif.
Nucleic Acids Res., 38, 2010
|
|
3M1M
 
 | | Crystal structure of the primase-polymerase from Sulfolobus islandicus | | Descriptor: | GLYCEROL, ORF904, SULFATE ION, ... | | Authors: | Vannini, A, Beck, K, Lipps, G, Cramer, P. | | Deposit date: | 2010-03-05 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The archaeo-eukaryotic primase of plasmid pRN1 requires a helix bundle domain for faithful primer synthesis
Nucleic Acids Res., 38, 2010
|
|
3NR5
 
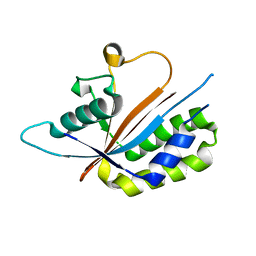 | | Crystal structure of human Maf1 | | Descriptor: | Repressor of RNA polymerase III transcription MAF1 homolog | | Authors: | Ringel, R, Vannini, A, Kusser, A.G, Berninghausen, O, Kassavetis, G.A, Cramer, P. | | Deposit date: | 2010-06-30 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular Basis of RNA Polymerase III Transcription Repression by Maf1
Cell(Cambridge,Mass.), 143, 2010
|
|
8RAN
 
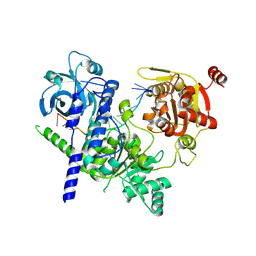 | |
8RAO
 
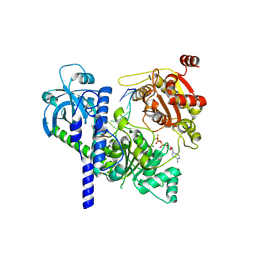 | |
8RBZ
 
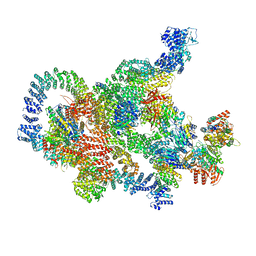 | | Structure of Integrator-PP2A-SOSS-CTD post-termination complex | | Descriptor: | DNA-directed RNA polymerase subunit, DSS1, Integrator complex subunit 1, ... | | Authors: | Fianu, I, Ochmann, M, Walshe, J.L, Cramer, P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of Integrator-dependent RNA polymerase II termination.
Nature, 629, 2024
|
|
2RF4
 
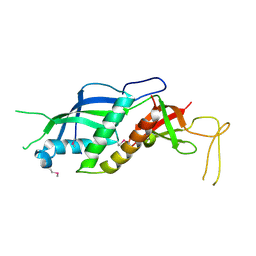 | |
2R93
 
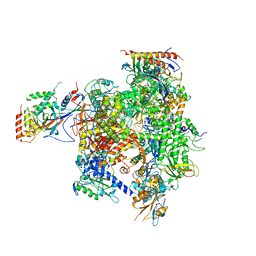 | | Elongation complex of RNA polymerase II with a hepatitis delta virus-derived RNA stem loop | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Lehmann, E, Brueckner, F, Cramer, P. | | Deposit date: | 2007-09-12 | | Release date: | 2007-11-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Molecular basis of RNA-dependent RNA polymerase II activity.
Nature, 450, 2007
|
|
8B3F
 
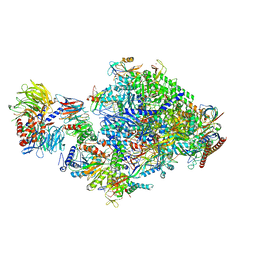 | | Pol II-CSB-CSA-DDB1-ELOF1 | | Descriptor: | DNA damage-binding protein 1, DNA excision repair protein ERCC-6, DNA excision repair protein ERCC-8, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Pol II-CSB-CSA-DDB1-ELOF1 structure.
To Be Published
|
|
