9D5A
 
 | |
6WZO
 
 | |
8CXK
 
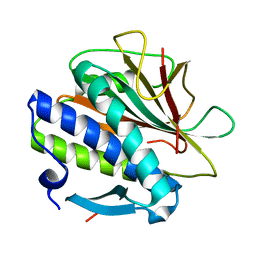 | | Structure of the C. elegans HIM-3 R93Y mutant | | Descriptor: | HORMA domain-containing protein | | Authors: | Ego, K.M, Russo, A, Giacopazzi, S, Deshong, A, Menon, M, Ortiz, V, Bhalla, N, Corbett, K.D. | | Deposit date: | 2022-05-21 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The conserved AAA ATPase PCH-2 distributes its regulation of meiotic prophase events through multiple meiotic HORMADs in C. elegans.
Plos Genet., 19, 2023
|
|
8CWW
 
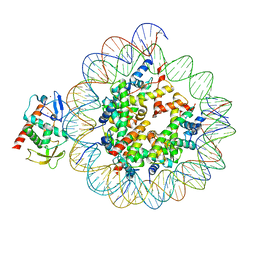 | | Structure of S. cerevisiae Hop1 CBR bound to a nucleosome | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Gu, Y, Ur, S.N, Milano, C.R, Tromer, E.C, Vale-Silva, L.A, Hochwagen, A, Corbett, K.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Chromatin binding by HORMAD proteins regulates meiotic recombination initiation.
Embo J., 43, 2024
|
|
8CZE
 
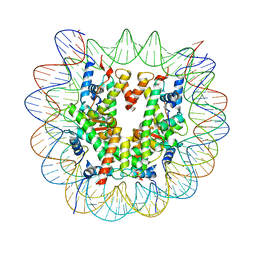 | | Structure of a Xenopus Nucleosome with Widom 601 DNA | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Gu, Y, Ur, S.N, Milano, C.R, Tromer, E.C, Vale-Silva, L.A, Hochwagen, A, Corbett, K.D. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Chromatin binding by HORMAD proteins regulates meiotic recombination initiation.
Embo J., 43, 2024
|
|
6DEI
 
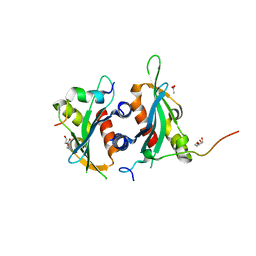 | | Structure of Dse3-Csm1 complex | | Descriptor: | ACETATE ION, Monopolin complex subunit CSM1, Protein DSE3, ... | | Authors: | Singh, N, Corbett, K.D. | | Deposit date: | 2018-05-12 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | The budding-yeast RWD protein Csm1 scaffolds diverse protein complexes through a conserved structural mechanism.
Protein Sci., 27, 2018
|
|
6DD8
 
 | | Structure of mouse SYCP3, P21 form | | Descriptor: | Synaptonemal complex protein 3 | | Authors: | Rosenberg, S.C, Munoz, I.C, Uson, I, Corbett, K.D. | | Deposit date: | 2018-05-09 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A conserved filamentous assembly underlies the structure of the meiotic chromosome axis.
Elife, 8, 2019
|
|
6DD9
 
 | | Structure of mouse SYCP3, P1 form | | Descriptor: | Synaptonemal complex protein 3 | | Authors: | Rosenberg, S.C, Munoz, I.C, Uson, I, Corbett, K.D. | | Deposit date: | 2018-05-09 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A conserved filamentous assembly underlies the structure of the meiotic chromosome axis.
Elife, 8, 2019
|
|
6DEX
 
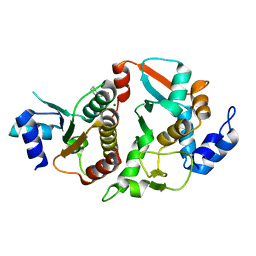 | | Structure of Eremothecium gossypii Shu1:Shu2 complex | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, MAGNESIUM ION, Suppressor of hydroxyurea sensitivity protein 1, ... | | Authors: | Singh, N, Corbett, K.D. | | Deposit date: | 2018-05-13 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Eremothecium gossypii Shu1:Shu2 complex
To Be Published
|
|
5CZO
 
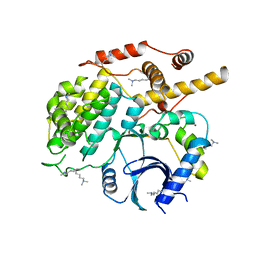 | | Structure of S. cerevisiae Hrr25:Mam1 complex, form 2 | | Descriptor: | Casein kinase I homolog HRR25, Monopolin complex subunit MAM1, ZINC ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
5CYA
 
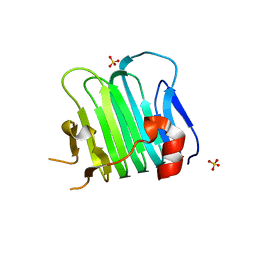 | | Crystal structure of Arl2 GTPase-activating protein tubulin cofactor C (TBCC) | | Descriptor: | SULFATE ION, Tubulin-specific chaperone C | | Authors: | Nithianantham, S, Le, S, Seto, E, Jia, W, Leary, J, Corbett, K.D, Moore, J.K, Al-Bassam, J. | | Deposit date: | 2015-07-30 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tubulin cofactors and Arl2 are cage-like chaperones that regulate the soluble alpha beta-tubulin pool for microtubule dynamics.
Elife, 4, 2015
|
|
5CYZ
 
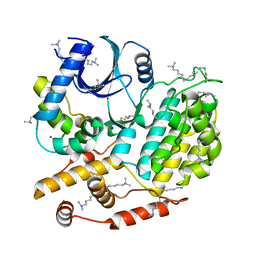 | | Structure of S. cerevisiae Hrr25:Mam1 complex, form 1 | | Descriptor: | Casein kinase I homolog HRR25, Monopolin complex subunit MAM1, ZINC ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
7UYX
 
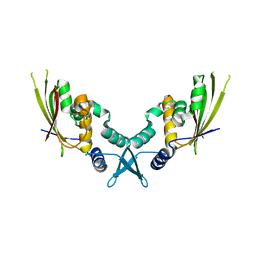 | | Structure of bacteriophage PA1c gp2 | | Descriptor: | Bacteriophage PA1C gp2 | | Authors: | Enustun, E, Deep, A, Gu, Y, Nguyen, K, Chaikeeratisak, V, Armbruster, E, Ghassemian, M, Pogliano, J, Corbett, K.D. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-10 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Identification of the bacteriophage nucleus protein interaction network.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3KAS
 
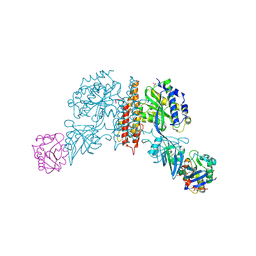 | | Machupo virus GP1 bound to human transferrin receptor 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, ... | | Authors: | Abraham, J, Corbett, K.D, Harrison, S.C. | | Deposit date: | 2009-10-19 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for receptor recognition by New World hemorrhagic fever arenaviruses.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6BZF
 
 | | Structure of S. cerevisiae Zip2:Spo16 complex, C2 form | | Descriptor: | Protein ZIP2, Sporulation-specific protein 16 | | Authors: | Arora, K, Corbett, K.D. | | Deposit date: | 2017-12-23 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.286 Å) | | Cite: | The conserved XPF:ERCC1-like Zip2:Spo16 complex controls meiotic crossover formation through structure-specific DNA binding.
Nucleic Acids Res., 47, 2019
|
|
6BZG
 
 | | Structure of S. cerevisiae Zip2:Spo16 complex, P212121 form | | Descriptor: | HEXAETHYLENE GLYCOL, Protein ZIP2, SULFATE ION, ... | | Authors: | Arora, K, Corbett, K.D. | | Deposit date: | 2017-12-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The conserved XPF:ERCC1-like Zip2:Spo16 complex controls meiotic crossover formation through structure-specific DNA binding.
Nucleic Acids Res., 47, 2019
|
|
4IM8
 
 | | low resolution crystal structure of mouse RAGE | | Descriptor: | Advanced glycation end-products receptor | | Authors: | Xu, D, Young, J.H, Krahn, J.M, Song, D, Corbett, K.D, Chazin, W.J, Pedersen, L.C, Esko, J.D. | | Deposit date: | 2013-01-02 | | Release date: | 2013-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Stable RAGE-Heparan Sulfate Complexes Are Essential for Signal Transduction.
Acs Chem.Biol., 8, 2013
|
|
7N0R
 
 | |
7N0I
 
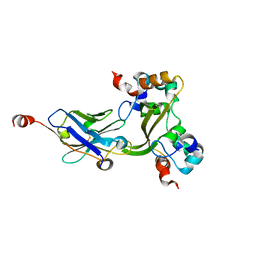 | |
2G2U
 
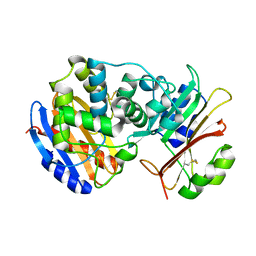 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Reynolds, K.A, Thomson, J.M, Corbett, K.D, Bethel, C.R, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2006-02-16 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Computational Characterization of the SHV-1 beta-Lactamase-beta-Lactamase Inhibitor Protein Interface.
J.Biol.Chem., 281, 2006
|
|
9C5G
 
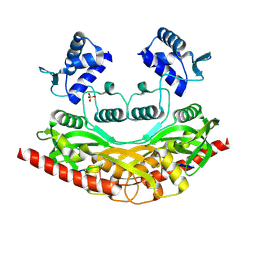 | |
2G2W
 
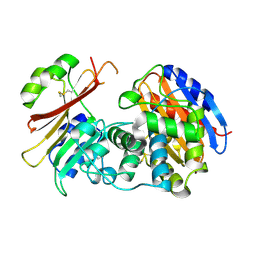 | | Crystal Structure of the SHV D104K Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein | | Authors: | Reynolds, K.A, Thomson, J.M, Corbett, K.D, Bethel, C.R, Berger, J.M, Kirsch, J.F, Bonomo, R.A, Handel, T.M. | | Deposit date: | 2006-02-16 | | Release date: | 2006-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Computational Characterization of the SHV-1 beta-Lactamase-beta-Lactamase Inhibitor Protein Interface.
J.Biol.Chem., 281, 2006
|
|
9CD2
 
 | |
7TQD
 
 | | Structure of Enterobacter cloacae Cap2-CdnD02 2:1 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gu, Y, Ye, Q, Ledvina, H.E, Quan, Y, Lau, R.K, Zhou, H, Whiteley, A.T, Corbett, K.D. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An E1-E2 fusion protein primes antiviral immune signalling in bacteria.
Nature, 616, 2023
|
|
8V47
 
 | | CryoEM structure of AriA-AriB complex (Form II) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AriA antitoxin, AriB | | Authors: | Deep, A, Corbett, K.D. | | Deposit date: | 2023-11-28 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Architecture and activation mechanism of the bacterial PARIS defence system.
Nature, 634, 2024
|
|
