8GFN
 
 | | Room temperature X-ray structure of truncated SARS-CoV-2 main protease C145A mutant, residues 1-304, in complex with BBH1 | | 分子名称: | (1R,2S,5S)-N-{(2S)-1-(1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Kovalevsky, A, Coates, L. | | 登録日 | 2023-03-08 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Contribution of the catalytic dyad of SARS-CoV-2 main protease to binding covalent and noncovalent inhibitors.
J.Biol.Chem., 299, 2023
|
|
8GFO
 
 | |
8GFR
 
 | | Room temperature X-ray structure of truncated SARS-CoV-2 main protease C145A mutant, residues 1-304, in complex with NBH2 | | 分子名称: | (1R,2S,5S)-N-{(1S)-1-cyano-2-[(3S)-2-oxopyrrolidin-3-yl]ethyl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Kovalevsky, A, Coates, L. | | 登録日 | 2023-03-08 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Contribution of the catalytic dyad of SARS-CoV-2 main protease to binding covalent and noncovalent inhibitors.
J.Biol.Chem., 299, 2023
|
|
7RMZ
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-63 | | 分子名称: | 3C-like proteinase, 6-{4-[3-chloro-4-(trifluoromethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-28 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RNK
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-71 | | 分子名称: | 3C-like proteinase, 6-{4-[3-chloro-4-(hydroxymethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(3H,5H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-29 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RME
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-52 | | 分子名称: | 3C-like proteinase, 6-{4-[4-chloro-3-(trifluoromethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-27 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RMB
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-78 | | 分子名称: | 3C-like proteinase, 6-[4-(4-bromo-3-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-27 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RN4
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-69 | | 分子名称: | 3C-like proteinase, 6-[4-(3,4-dichlorophenyl)piperidine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-29 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RM2
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule-CSR-494190-S1 | | 分子名称: | 3C-like proteinase, 6-[4-(3,5-dichloro-4-methylphenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-26 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RNH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-45 | | 分子名称: | 3C-like proteinase, 6-[4-(4-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-29 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RMT
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-70 | | 分子名称: | 2-chloro-4-[4-(2,6-dioxo-1,2,5,6-tetrahydropyrimidine-4-carbonyl)piperazin-1-yl]benzaldehyde, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-28 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RLS
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-68 | | 分子名称: | 3C-like proteinase, 6-[4-(3,4,5-trichlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-26 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7JY2
 
 | | Z-DNA joint X-ray/Neutron | | 分子名称: | Chains: A,B | | 著者 | Harp, J.M, Coates, L, Egli, M. | | 登録日 | 2020-08-28 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | 主引用文献 | Water structure around a left-handed Z-DNA fragment analyzed by cryo neutron crystallography.
Nucleic Acids Res., 49, 2021
|
|
5A93
 
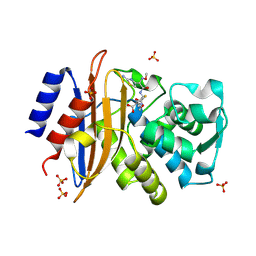 | | 293K Joint X-ray Neutron with Cefotaxime: EXPLORING THE MECHANISM OF BETA-LACTAM RING PROTONATION IN THE CLASS A BETA-LACTAMASE ACYLATION MECHANISM USING NEUTRON AND X-RAY CRYSTALLOGRAPHY | | 分子名称: | BETA-LACTAMASE CTX-M-97, CEFOTAXIME, C3' cleaved, ... | | 著者 | Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Erskine, P.T, Tomanicek, S.J, Ostermann, A, Schrader, T.E, Ginell, S.L, Coates, L. | | 登録日 | 2015-07-17 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-01-10 | | 実験手法 | NEUTRON DIFFRACTION (1.598 Å), X-RAY DIFFRACTION | | 主引用文献 | Exploring the Mechanism of Beta-Lactam Ring Protonation in the Class a Beta-Lactamase Acylation Mechanism Using Neutron and X-Ray Crystallography.
J.Med.Chem., 59, 2016
|
|
5A90
 
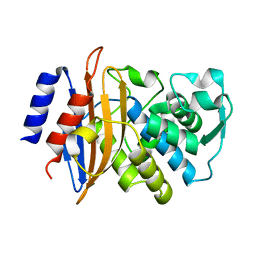 | | 100K Neutron Ligand Free: Exploring the Mechanism of beta-Lactam Ring Protonation in the Class A beta-lactamase Acylation Mechanism Using Neutron and X-ray Crystallography | | 分子名称: | BETA-LACTAMASE CTX-M-97 | | 著者 | Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Erskine, P.T, Tomanicek, S.J, Ostermann, A, Schrader, T.E, Ginell, S.L, Coates, L. | | 登録日 | 2015-07-17 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | NEUTRON DIFFRACTION (1.7 Å) | | 主引用文献 | Exploring the Mechanism of Beta-Lactam Ring Protonation in the Class a Beta-Lactamase Acylation Mechanism Using Neutron and X-Ray Crystallography.
J.Med.Chem., 59, 2016
|
|
5A91
 
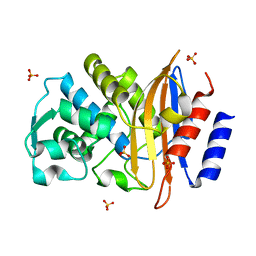 | | 15K X-ray ligand free: Exploring the Mechanism of beta-Lactam Ring Protonation in the Class A beta-lactamase Acylation Mechanism Using Neutron and X-ray Crystallography | | 分子名称: | Beta-lactamase Toho-1, SULFATE ION | | 著者 | Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Erskine, P.T, Tomanicek, S.J, Ostermann, A, Schrader, T.E, Ginell, S.L, Coates, L. | | 登録日 | 2015-07-17 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Exploring the Mechanism of Beta-Lactam Ring Protonation in the Class a Beta-Lactamase Acylation Mechanism Using Neutron and X-Ray Crystallography.
J.Med.Chem., 59, 2016
|
|
5A92
 
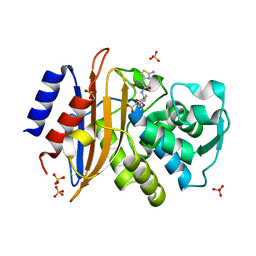 | | 15K X-ray structure with Cefotaxime: Exploring the Mechanism of beta- Lactam Ring Protonation in the Class A beta-lactamase Acylation Mechanism Using Neutron and X-ray Crystallography | | 分子名称: | BETA-LACTAMASE CTX-M-97, CEFOTAXIME, C3' cleaved, ... | | 著者 | Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Erskine, P.T, Tomanicek, S.J, Ostermann, A, Schrader, T.E, Ginell, S.L, Coates, L. | | 登録日 | 2015-07-17 | | 公開日 | 2015-12-16 | | 最終更新日 | 2018-10-03 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Exploring the Mechanism of Beta-Lactam Ring Protonation in the Class a Beta-Lactamase Acylation Mechanism Using Neutron and X-Ray Crystallography.
J.Med.Chem., 59, 2016
|
|
8GFU
 
 | |
6C78
 
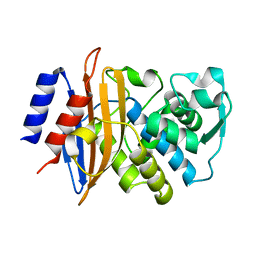 | | Substrate Binding Induces Conformational Changes In A Class A Beta Lactamase That Primes It For Catalysis | | 分子名称: | Beta-lactamase Toho-1 | | 著者 | Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M, Coates, L. | | 登録日 | 2018-01-22 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | NEUTRON DIFFRACTION (1.75 Å) | | 主引用文献 | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
7UKK
 
 | |
7UJG
 
 | |
7UJU
 
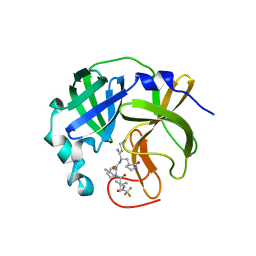 | | Room-temperature X-ray structure of monomeric SARS-CoV-2 main protease catalytic domain (MPro1-196) in complex with nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2022-03-31 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Autoprocessing and oxyanion loop reorganization upon GC373 and nirmatrelvir binding of monomeric SARS-CoV-2 main protease catalytic domain.
Commun Biol, 5, 2022
|
|
7KKU
 
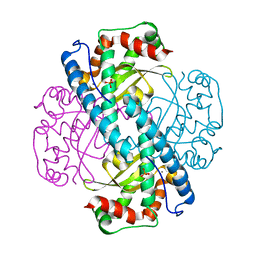 | | X-ray Counterpart to Neutron Structure of Oxidized Human MnSOD | | 分子名称: | MANGANESE (III) ION, PHOSPHATE ION, Superoxide dismutase [Mn], ... | | 著者 | Azadmanesh, J, Lutz, W.E, Coates, L, Weiss, K.L, Borgstahl, G.E.O. | | 登録日 | 2020-10-28 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Direct detection of coupled proton and electron transfers in human manganese superoxide dismutase.
Nat Commun, 12, 2021
|
|
7KLB
 
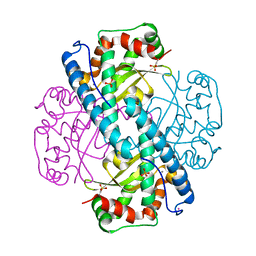 | | X-ray Counterpart to Neutron Structure of Reduced Human MnSOD | | 分子名称: | MANGANESE (II) ION, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Azadmanesh, J, Lutz, W.E, Coates, L, Weiss, K.L, Borgstahl, G.E.O. | | 登録日 | 2020-10-29 | | 公開日 | 2021-04-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Direct detection of coupled proton and electron transfers in human manganese superoxide dismutase.
Nat Commun, 12, 2021
|
|
7KKW
 
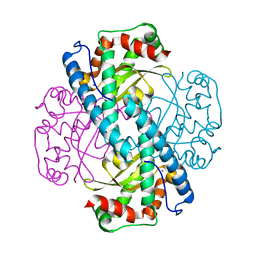 | | Neutron structure of Reduced Human MnSOD | | 分子名称: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial, ... | | 著者 | Azadmanesh, J, Lutz, W.E, Coates, L, Weiss, K.L, Borgstahl, G.E.O. | | 登録日 | 2020-10-28 | | 公開日 | 2021-04-21 | | 最終更新日 | 2024-04-10 | | 実験手法 | NEUTRON DIFFRACTION (2.3 Å) | | 主引用文献 | Direct detection of coupled proton and electron transfers in human manganese superoxide dismutase.
Nat Commun, 12, 2021
|
|
