5KC8
 
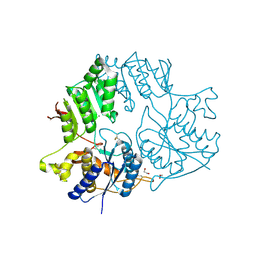 | | Crystal structure of the amino-terminal domain (ATD) of iGluR Delta-2 (GluD2) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glutamate receptor ionotropic, ... | | Authors: | Elegheert, J, Clay, J.E, Siebold, C, Aricescu, A.R. | | Deposit date: | 2016-06-05 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structural basis for integration of GluD receptors within synaptic organizer complexes.
Science, 353, 2016
|
|
5KC5
 
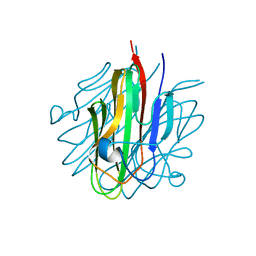 | | Crystal structure of the Cbln1 C1q domain trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Elegheert, J, Clay, J.E, Siebold, C, Aricescu, A.R. | | Deposit date: | 2016-06-05 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structural basis for integration of GluD receptors within synaptic organizer complexes.
Science, 353, 2016
|
|
5KC9
 
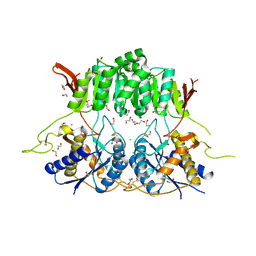 | | Crystal structure of the amino-terminal domain (ATD) of iGluR Delta-1 (GluD1) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Elegheert, J, Clay, J.E, Siebold, C, Aricescu, A.R. | | Deposit date: | 2016-06-05 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for integration of GluD receptors within synaptic organizer complexes.
Science, 353, 2016
|
|
5KC6
 
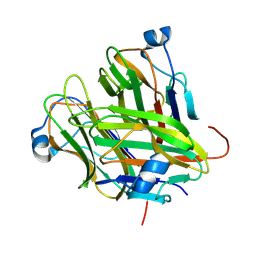 | |
5LGW
 
 | | GlgE isoform 1 from Streptomyces coelicolor D394A mutant co-crystallised with maltodextrin | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, CITRIC ACID, ... | | Authors: | Syson, K, Stevenson, C.E.M, Mia, F, Barclay, J.E, Tang, M, Gorelik, A, Rashid, A.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2016-07-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
5LGV
 
 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant soaked in maltooctaose | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Mia, F, Barclay, J.E, Tang, M, Gorelik, A, Rashid, A.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2016-07-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
