1KT1
 
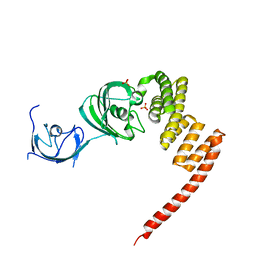 | | Structure of the Large FKBP-like Protein, FKBP51, Involved in Steroid Receptor Complexes | | Descriptor: | FK506-binding protein FKBP51, SULFATE ION | | Authors: | Sinars, C.R, Cheung-Flynn, J, Rimerman, R.A, Scammell, J.G, Smith, D.F, Clardy, J.C. | | Deposit date: | 2002-01-14 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | STRUCTURE OF THE LARGE FK506-BINDING PROTEIN FKBP51, AN HSP90-BINDING PROTEIN AND A COMPONENT OF STEROID RECEPTOR COMPLEXES
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1KT0
 
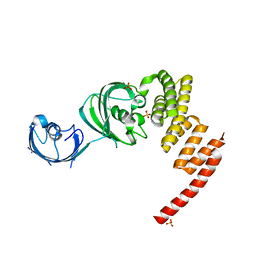 | | Structure of the Large FKBP-like Protein, FKBP51, Involved in Steroid Receptor Complexes | | Descriptor: | 51 KDA FK506-BINDING PROTEIN, SULFATE ION | | Authors: | Sinars, C.R, Cheung-Flynn, J, Rimerman, R.A, Scammell, J.G, Smith, D.F, Clardy, J.C. | | Deposit date: | 2002-01-14 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the large FK506-binding protein FKBP51, an Hsp90-binding protein and a
component of steroid receptor complexes
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1GRN
 
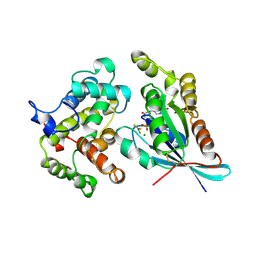 | | CRYSTAL STRUCTURE OF THE CDC42/CDC42GAP/ALF3 COMPLEX. | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Nassar, N, Hoffman, G.R, Clardy, J.C, Cerione, R.A. | | Deposit date: | 1998-07-30 | | Release date: | 1999-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP.
Nat.Struct.Biol., 5, 1998
|
|
1BN5
 
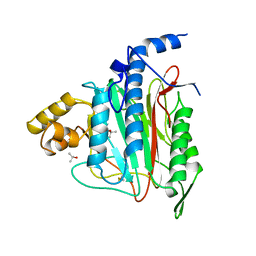 | | HUMAN METHIONINE AMINOPEPTIDASE 2 | | Descriptor: | COBALT (II) ION, METHIONINE AMINOPEPTIDASE, TERTIARY-BUTYL ALCOHOL | | Authors: | Liu, S, Widom, J, Kemp, C.W, Crews, C.M, Clardy, J.C. | | Deposit date: | 1998-07-31 | | Release date: | 1999-07-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
1BOA
 
 | | HUMAN METHIONINE AMINOPEPTIDASE 2 COMPLEXED WITH ANGIOGENESIS INHIBITOR FUMAGILLIN | | Descriptor: | COBALT (II) ION, FUMAGILLIN, METHIONINE AMINOPEPTIDASE | | Authors: | Liu, S, Widom, J, Kemp, C.W, Crews, C.M, Clardy, J.C. | | Deposit date: | 1998-08-01 | | Release date: | 1999-08-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
1B6A
 
 | | HUMAN METHIONINE AMINOPEPTIDASE 2 COMPLEXED WITH TNP-470 | | Descriptor: | (1R,2S,3S,4R)-4-hydroxy-2-methoxy-4-methyl-3-[(2R,3R)-2-methyl-3-(3-methylbut-2-en-1-yl)oxiran-2-yl]cyclohexyl (chloroacetyl)carbamate, COBALT (II) ION, METHIONINE AMINOPEPTIDASE | | Authors: | Liu, S, Clardy, J.C. | | Deposit date: | 1999-01-13 | | Release date: | 2000-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
1B59
 
 | | COMPLEX OF HUMAN METHIONINE AMINOPEPTIDASE-2 COMPLEXED WITH OVALICIN | | Descriptor: | 3,4-DIHYDROXY-2-METHOXY-4-METHYL-3-[2-METHYL-3-(3-METHYL-BUT-2-ENYL) -OXIRANYL]-CYCLOHEXANONE, COBALT (II) ION, PROTEIN (METHIONINE AMINOPEPTIDASE) | | Authors: | Liu, S, Clardy, J.C. | | Deposit date: | 1999-01-13 | | Release date: | 2000-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human methionine aminopeptidase-2 complexed with fumagillin.
Science, 282, 1998
|
|
1FKB
 
 | | ATOMIC STRUCTURE OF THE RAPAMYCIN HUMAN IMMUNOPHILIN FKBP-12 COMPLEX | | Descriptor: | FK506 BINDING PROTEIN, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Van Duyne, G.D, Standaert, R.F, Schreiber, S.L, Clardy, J.C. | | Deposit date: | 1992-07-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atomic Structure of the Rapamycin Human Immunophilin Fkbp-12 Complex
J.Am.Chem.Soc., 113, 1991
|
|
