3CED
 
 | | Crystal structure of the C-terminal NIL domain of an ABC transporter protein homologue from Staphylococcus aureus | | Descriptor: | 1,4-BUTANEDIOL, Methionine import ATP-binding protein metN 2 | | Authors: | Cuff, M.E, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-28 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of the C-terminal NIL domain of an ABC transporter protein homologue from Staphylococcus aureus.
TO BE PUBLISHED
|
|
3M6Y
 
 | | Structure of 4-hydroxy-2-oxoglutarate aldolase from bacillus cereus at 1.45 a resolution. | | Descriptor: | 4-Hydroxy-2-oxoglutarate aldolase, CALCIUM ION, CHLORIDE ION | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-16 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of 4-Hydroxy-2-Oxoglutarate Aldolase from Bacillus Cereus at 1.45 A Resolution.
To be Published
|
|
4P7C
 
 | | Crystal structure of putative methyltransferase from Pseudomonas syringae pv. tomato | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, tRNA (mo5U34)-methyltransferase | | Authors: | Chang, C, Mack, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of putative methyltransferase from Pseudomonas syringae pv. tomato
To Be Published
|
|
4MQD
 
 | | Crystal structure of ComJ, inhibitor of the DNA degrading activity of NucA, from Bacillus subtilis | | Descriptor: | DNA-entry nuclease inhibitor | | Authors: | Chang, C, Mack, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-16 | | Release date: | 2013-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of ComJ, inhibitor of the DNA degrading activity of NucA, from Bacillus subtilis
To be Published
|
|
4OVJ
 
 | |
4R9N
 
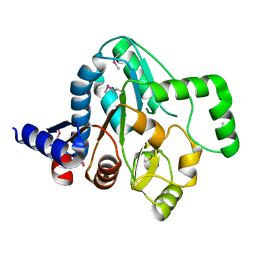 | |
4ESY
 
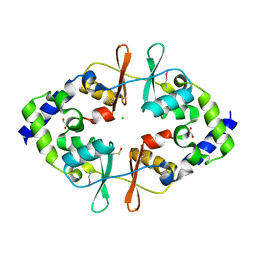 | | Crystal Structure of the CBS Domain of CBS Domain Containing Membrane Protein from Sphaerobacter thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CBS domain containing membrane protein, CHLORIDE ION | | Authors: | Kim, Y, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-09-05 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal Structure of the CBS Domain of CBS Domain Containing Membrane Protein from Sphaerobacter thermophilus
To be Published
|
|
3BV8
 
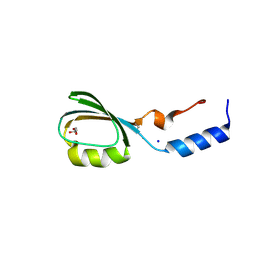 | | Crystal structure of the N-terminal domain of tetrahydrodipicolinate acetyltransferase from Staphylococcus aureus | | Descriptor: | GLYCEROL, SODIUM ION, Tetrahydrodipicolinate acetyltransferase | | Authors: | Cuff, M.E, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the N-terminal domain of tetrahydrodipicolinate acetyltransferase from Staphylococcus aureus.
TO BE PUBLISHED
|
|
3OJ0
 
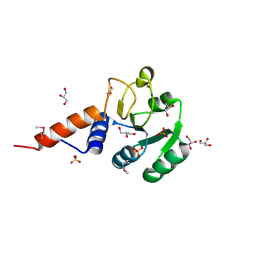 | | Crystal structure of glutamyl-tRNA reductase from Thermoplasma volcanium (nucleotide binding domain) | | Descriptor: | GLYCEROL, Glutamyl-tRNA reductase, SULFATE ION | | Authors: | Michalska, K, Marshall, N, Clancy, S, Puttagunta, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-20 | | Release date: | 2010-09-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Crystal structure of glutamyl-tRNA reductase from Thermoplasma volcanium (nucleotide binding domain)
To be Published
|
|
3BZ6
 
 | |
3OOO
 
 | | The structure of a proline dipeptidase from Streptococcus agalactiae 2603V | | Descriptor: | Proline dipeptidase | | Authors: | Fan, Y, Wu, R, Morales, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The structure of a proline dipeptidase from Streptococcus agalactiae 2603V
To be Published
|
|
3EA0
 
 | | Crystal Structure of ParA Family ATPase from Chlorobium tepidum TLS | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATPase, ParA family, ... | | Authors: | Kim, Y, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-24 | | Release date: | 2008-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of ParA Family ATPase from Chlorobium tepidum TLS
To be Published
|
|
3EAG
 
 | |
3PFM
 
 | | Crystal structure of a EAL domain of GGDEF domain protein from Pseudomonas fluorescens Pf | | Descriptor: | GGDEF domain protein | | Authors: | Nocek, B, Stein, A, Marshall, N, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-10-28 | | Release date: | 2010-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Crystal structure of a EAL domain of GGDEF domain protein from Pseudomonas fluorescens Pf
TO BE PUBLISHED
|
|
3CCY
 
 | |
3PU9
 
 | | Crystal structure of serine/threonine phosphatase Sphaerobacter thermophilus DSM 20745 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Protein serine/threonine phosphatase | | Authors: | Nocek, B, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of serine/threonine phosphatase Sphaerobacter thermophilus DSM 20745
TO BE PUBLISHED
|
|
3PN9
 
 | |
3CLQ
 
 | |
3QZ6
 
 | |
4Q7Q
 
 | | The crystal structure of a possible lipase from Chitinophaga pinensis DSM 2588 | | Descriptor: | CHLORIDE ION, FORMIC ACID, Lipolytic protein G-D-S-L family, ... | | Authors: | Tan, K, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | The crystal structure of a possible lipase from Chitinophaga pinensis DSM 2588
To be Published
|
|
3QQZ
 
 | | Crystal structure of the C-terminal domain of the yjiK protein from Escherichia coli CFT073 | | Descriptor: | CALCIUM ION, Putative uncharacterized protein yjiK | | Authors: | Stein, A, Chhor, G, Nocek, B, Fenske, R.J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-16 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the C-terminal domain of the yjiK protein from Escherichia coli CFT073
TO BE PUBLISHED
|
|
3EXN
 
 | | Crystal structure of acetyltransferase from Thermus thermophilus HB8 | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, Probable acetyltransferase | | Authors: | Nocek, B, Hatzos, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of acetyltransferase from Thermus thermophilus HB8
To be Published
|
|
3NE7
 
 | | Crystal structure of paia n-acetyltransferase from thermoplasma acidophilum in complex with coenzyme a | | Descriptor: | ACETYLTRANSFERASE, BETA-MERCAPTOETHANOL, COENZYME A, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
4MTN
 
 | |
3OOV
 
 | | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287 | | Descriptor: | GLYCEROL, Methyl-accepting chemotaxis protein, putative | | Authors: | Joachimiak, A, Duke, N.E.C, Hatzos-Skintges, C, Mulligan, R, Clancy, S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-08 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a methyl-accepting chemotaxis protein, residues 122 to 287
To be Published
|
|
