7ZSF
 
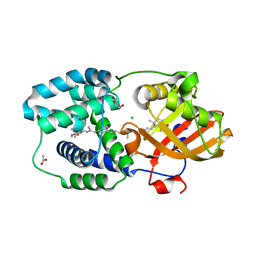 | | Structure of Orange Carotenoid Protein with canthaxanthin bound | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
7ZSJ
 
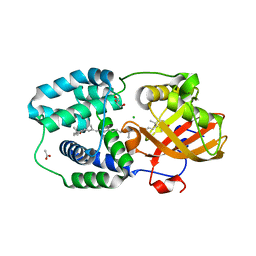 | | Structure of Orange Carotenoid Protein with canthaxanthin bound after 10 minutes of illumination | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
7ZSG
 
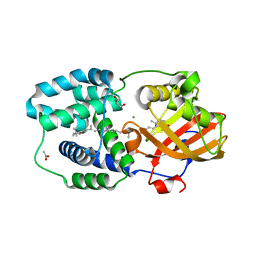 | | Structure of Orange Carotenoid Protein with canthaxanthin bound after 1 minute of illumination | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chukhutsina, V.U, Baxter, J.M, Fadini, A, Morgan, R.M, Pope, M.A, Maghlaoui, K, Orr, C, Wagner, A, van Thor, J.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Light activation of Orange Carotenoid Protein reveals bicycle-pedal single-bond isomerization.
Nat Commun, 13, 2022
|
|
1LCC
 
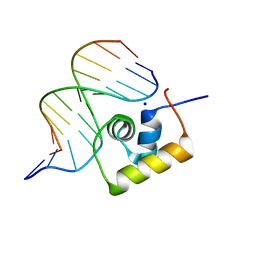 | | STRUCTURE OF THE COMPLEX OF LAC REPRESSOR HEADPIECE AND AN 11 BASE-PAIR HALF-OPERATOR DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), Lac Repressor, ... | | Authors: | Chuprina, V.P, Rullmann, J.A.C, Lamerichs, R.M.J.N, Van Boom, J.H, Boelens, R, Kaptein, R. | | Deposit date: | 1993-03-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 234, 1993
|
|
1LCD
 
 | | STRUCTURE OF THE COMPLEX OF LAC REPRESSOR HEADPIECE AND AN 11 BASE-PAIR HALF-OPERATOR DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), Lac Repressor, ... | | Authors: | Chuprina, V.P, Rullmann, J.A.C, Lamerichs, R.M.J.N, Van Boom, J.H, Boelens, R, Kaptein, R. | | Deposit date: | 1993-03-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 234, 1993
|
|
1KBU
 
 | | CRE RECOMBINASE BOUND TO A LOXP HOLLIDAY JUNCTION | | Descriptor: | CRE RECOMBINASE, LOXP | | Authors: | Martin, S.S, Pulido, E, Chu, V.C, Lechner, T, Baldwin, E.P. | | Deposit date: | 2001-11-06 | | Release date: | 2002-06-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Order of Strand Exchanges in Cre-LoxP Recombination and its Basis Suggested by the Crystal Structure of a
Cre-LoxP Holliday Junction Complex
J.Mol.Biol., 319, 2002
|
|
1MA7
 
 | |
4GDA
 
 | | Circular Permuted Streptavidin A50/N49 | | Descriptor: | BIOTIN, ETHANOL, GLYCEROL, ... | | Authors: | Le Trong, I, Chu, V, Xing, Y, Lybrand, T.P, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2012-07-31 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural consequences of cutting a binding loop: two circularly permuted variants of streptavidin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1SWG
 
 | | CIRCULAR PERMUTED STREPTAVIDIN E51/A46 IN COMPLEX WITH BIOTIN | | Descriptor: | BIOTIN, CIRCULARLY PERMUTED CORE-STREPTAVIDIN E51/A46 | | Authors: | Freitag, S, Chu, V, Le Trong, I, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-07-12 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamic and structural consequences of flexible loop deletion by circular permutation in the streptavidin-biotin system.
Protein Sci., 7, 1998
|
|
1SWF
 
 | | CIRCULAR PERMUTED STREPTAVIDIN E51/A46 | | Descriptor: | CIRCULARLY PERMUTED CORE-STREPTAVIDIN E51/A46 | | Authors: | Freitag, S, Chu, V, Le Trong, I, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1997-04-23 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural consequences of flexible loop deletion by circular permutation in the streptavidin-biotin system.
Protein Sci., 7, 1998
|
|
4GD9
 
 | | Circular Permuted Streptavidin N49/G48 | | Descriptor: | BIOTIN, SULFATE ION, Streptavidin | | Authors: | Le Trong, I, Chu, V, Xing, Y, Lybrand, T.P, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2012-07-31 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural consequences of cutting a binding loop: two circularly permuted variants of streptavidin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1N9Y
 
 | | Streptavidin Mutant S27A at 1.5A Resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-11-26 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1N9M
 
 | | Streptavidin Mutant S27A with Biotin at 1.6A Resolution | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-11-25 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1N7Y
 
 | | STREPTAVIDIN MUTANT N23E AT 1.96A | | Descriptor: | Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-11-18 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1N43
 
 | | Streptavidin Mutant N23A with biotin at 1.89A | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-10-30 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1N4J
 
 | | STREPTAVIDIN MUTANT N23A AT 2.18A | | Descriptor: | Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-10-31 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NC9
 
 | | STREPTAVIDIN MUTANT Y43A WITH IMINOBIOTIN AT 1.8A RESOLUTION | | Descriptor: | 2-IMINOBIOTIN, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-12-05 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NDJ
 
 | | Streptavidin Mutant Y43F with Biotin at 1.81A Resolution | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-12-09 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1SWT
 
 | | CORE-STREPTAVIDIN MUTANT D128A IN COMPLEX WITH BIOTIN AT PH 4.5 | | Descriptor: | BIOTIN, PROTEIN (STREPTAVIDIN) | | Authors: | Freitag, S, Le Trong, I, Chu, V, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-10-22 | | Release date: | 1999-07-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural snapshot of an intermediate on the streptavidin-biotin dissociation pathway.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1SWS
 
 | | CORE-STREPTAVIDIN MUTANT D128A AT PH 4.5 | | Descriptor: | PROTEIN (STREPTAVIDIN) | | Authors: | Freitag, S, Chu, V, Le Trong, I, Klumb, L.A, To, R, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-10-22 | | Release date: | 1999-07-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural snapshot of an intermediate on the streptavidin-biotin dissociation pathway.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1NBX
 
 | | Streptavidin Mutant Y43A at 1.70A Resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Streptavidin | | Authors: | Le Trong, I, Freitag, S, Klumb, L.A, Chu, V, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 2002-12-04 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of hydrogen bonds in the high-affinity streptavidin-biotin complex: mutations of amino acids interacting with the ureido oxygen of biotin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2KS1
 
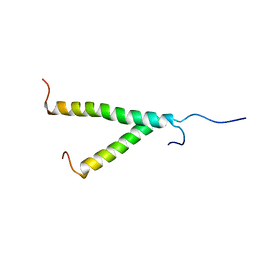 | | Heterodimeric association of Transmembrane domains of ErbB1 and ErbB2 receptors Enabling Kinase Activation | | Descriptor: | Epidermal growth factor receptor, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Mineev, K.S, Bocharov, E.V, Pustovalova, Y.E, Bocharova, O.V, Chupin, V.V, Arseniev, A.S. | | Deposit date: | 2009-12-24 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of the Transmembrane Domain Heterodimer of ErbB1 and ErbB2 Receptor Tyrosine Kinases
J.Mol.Biol., 2010
|
|
6T64
 
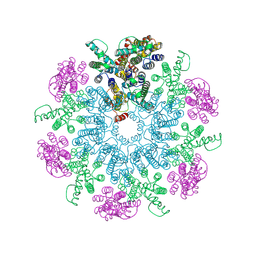 | | A model of the EIAV CA-SP hexamer (C6) from Gag-deltaMA spheres assembled at pH6 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
6T63
 
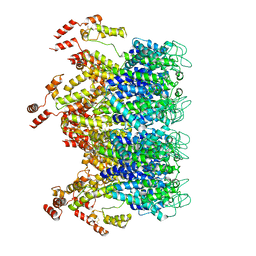 | | A model of the EIAV CA-SP hexamer (C2) from Gag-deltaMA tubes assembled at pH6 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
6T61
 
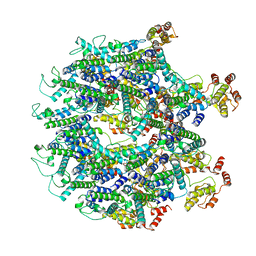 | | A model of the EIAV CA-SP hexamer (C2) from Gag-deltaMA tubes assembled at pH8 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
