6BYK
 
 | | Structure of 14-3-3 beta/alpha bound to O-ClcNAc peptide | | Descriptor: | 14-3-3 protein beta/alpha, 2-acetamido-2-deoxy-beta-D-glucopyranose, ATPPVSQASSTT O-GlcNac peptide | | Authors: | Schumacher, M.A. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
6CHV
 
 | | Proteus vulgaris HigA antitoxin bound to DNA | | Descriptor: | Antitoxin HigA, MAGNESIUM ION, pHigCryst3, ... | | Authors: | Schureck, M.A, Hoffer, E.D, Onuoha, N, Dunham, C.M. | | Deposit date: | 2018-02-23 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcriptional regulation by the HigA antitoxin.
Mol.Microbiol., 111, 2019
|
|
8TFK
 
 | |
8TGE
 
 | |
8TFB
 
 | |
6DXO
 
 | | 1.8 A structure of RsbN-BldN complex. | | Descriptor: | BldN, RNA polymerase ECF-subfamily sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-06-29 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the RsbN-sigma BldN complex from Streptomyces venezuelae defines a new structural class of anti-sigma factor.
Nucleic Acids Res., 46, 2018
|
|
7TZV
 
 | | Structure of DriD C-domain bound to 9mer ssDNA | | Descriptor: | DNA (5'-D(*TP*AP*GP*TP*CP*TP*AP*CP*T)-3'), WYL domain-containing protein | | Authors: | Schumacher, M.A, Laub, M. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | ssDNA is an allosteric regulator of the C. crescentus SOS-independent DNA damage response transcription activator, DriD.
Genes Dev., 36, 2022
|
|
6BZD
 
 | |
5FQL
 
 | | Insights into Hunter syndrome from the structure of iduronate-2- sulfatase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Demydchuk, M, Hill, C.H, Zhou, A, Bunkoczi, G, Stein, P.E, Marchesan, D, Deane, J.E, Read, R.J. | | Deposit date: | 2015-12-11 | | Release date: | 2017-01-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into Hunter syndrome from the structure of iduronate-2-sulfatase.
Nat Commun, 8, 2017
|
|
5KEC
 
 | | Structure of K. pneumonia MrkH in its apo state. | | Descriptor: | Flagellar brake protein YcgR | | Authors: | Schumacher, M. | | Deposit date: | 2016-06-09 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | to be published:
Structures of K. pneumonia MrkH: dual utilization of the PilZ fold for c-di-GMP and DNA binding by a novel activator of biofilm genes
To Be Published
|
|
5KGO
 
 | | Structure of K. pneumonia MrkH-c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Flagellar brake protein YcgR | | Authors: | Schumacher, M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | To be published:
Structures of K. pneumonia MrkH: dual utilization of the PilZ fold for c-di-GMP and DNA binding by a novel activator of biofilm genes
To Be Published
|
|
8BAR
 
 | |
8BAS
 
 | | E. coli C7 DarT1 in complex with carba-NAD and DNA | | Descriptor: | 1,2-ETHANEDIOL, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, DNA (5'-D(*AP*AP*GP*AP*C)-3'), ... | | Authors: | Schuller, M, Ariza, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Molecular basis for the reversible ADP-ribosylation of guanosine bases.
Mol.Cell, 83, 2023
|
|
5HBU
 
 | |
6CG8
 
 | | Structure of C. crescentus GapR-DNA | | Descriptor: | DNA (5'-D(*TP*TP*AP*AP*AP*AP*TP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*AP*TP*TP*TP*TP*AP*A)-3'), UPF0335 protein B7Z12_12435 | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-02-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication.
Cell, 175, 2018
|
|
8P3E
 
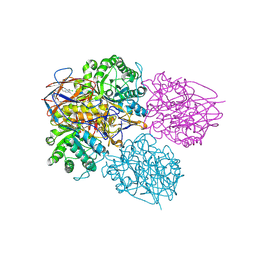 | | Crystal structure of glucocerebrosidase in complex with allosteric activator | | Descriptor: | 2-[[3-[(4-chlorophenyl)carbamoyl]phenyl]sulfonylamino]benzoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schulze, M.-S. | | Deposit date: | 2023-05-17 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of ss-Glucocerebrosidase Activators for Glucosylceramide hydrolysis.
Chemmedchem, 19, 2024
|
|
5KED
 
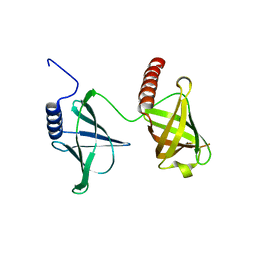 | | Structure of the 2.65 Angstrom P2(1) crystal of K. pneumonia MrkH | | Descriptor: | Flagellar brake protein YcgR | | Authors: | Schumacher, M. | | Deposit date: | 2016-06-09 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | To be released:
Structures of K. pneumonia MrkH: dual utilization of the PilZ fold for c-di-GMP and DNA binding by a novel activator of biofilm genes
To Be Published
|
|
6CFX
 
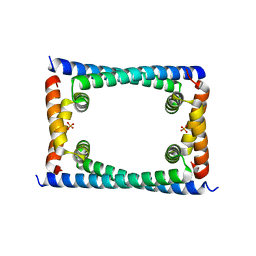 | | Bosea sp GapR solved in the presence of DNA | | Descriptor: | PHOSPHATE ION, UPF0335 protein ASE63_04290 | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-02-18 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication.
Cell, 175, 2018
|
|
8SUK
 
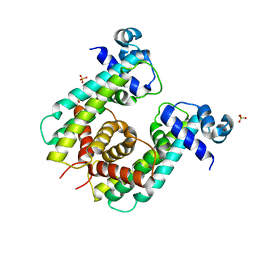 | | Structure of Rhodococcus sp. USK13 DarR-c-di-AMP complex | | Descriptor: | DNA (5'-D(*AP*A)-3'), DarR, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-12 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
8SVA
 
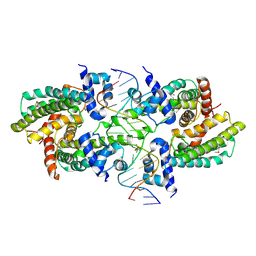 | | Structure of the Rhodococcus sp. USK13 DarR-20 bp DNA complex | | Descriptor: | DNA (5'-D(*TP*AP*GP*AP*TP*AP*CP*TP*CP*CP*GP*GP*AP*GP*TP*AP*TP*CP*TP*A)-3'), PHOSPHATE ION, TetR/AcrR family transcriptional regulator | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
8SV6
 
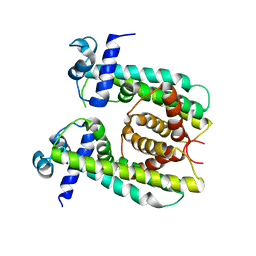 | | Structure of the M. smegmatis DarR protein | | Descriptor: | Fatty acid metabolism regulator protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
8SUA
 
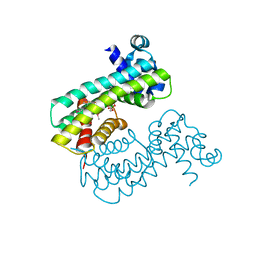 | | Structure of M. baixiangningiae DarR-ligand complex | | Descriptor: | 3-azanyl-3-(hydroxymethyl)-1,5,7,11-tetraoxa-6$l^{4}-boraspiro[5.5]undecan-9-ol, DarR | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
6YB5
 
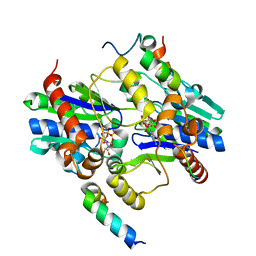 | | Orthorhombic crystal structure of a native BcsRQ complex crystallized in the presence of ADP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bacterial cellulose secretion regulator BcsQ, Bacterial cellulose secretion regulator BcsR, ... | | Authors: | Caleechurn, M, Abidi, W, Zouhir, S, Roche, S, Krasteva, P.V. | | Deposit date: | 2020-03-15 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Architecture and regulation of an enterobacterial cellulose secretion system.
Sci Adv, 7, 2021
|
|
8CDG
 
 | | Crystal structure of human IL-17A cytokine in complex with macrocycle | | Descriptor: | (9~{S},12~{R},19~{S})-9-[[4-[[(2~{S})-2-cyclohexyl-2-(2-phenylethanoylamino)ethanoyl]amino]phenyl]methyl]-12-methyl-7,10,13,21-tetrakis(oxidanylidene)-8,11,14,20-tetrazaspiro[4.17]docosane-19-carboxylic acid, Interleukin-17A | | Authors: | Schulze, M.-S. | | Deposit date: | 2023-01-30 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Modulation of IL-17 backbone dynamics reduces receptor affinity and reveals a new inhibitory mechanism.
Chem Sci, 14, 2023
|
|
6CF1
 
 | | Proteus vulgaris HigA antitoxin structure | | Descriptor: | Antitoxin HigA, POTASSIUM ION | | Authors: | Schureck, M.A, Hoffer, E.D, Ei Cho, S, Dunham, C.M. | | Deposit date: | 2018-02-13 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of transcriptional regulation by the HigA antitoxin.
Mol.Microbiol., 111, 2019
|
|
