5BQL
 
 | | Fluorescent protein cyOFP | | Descriptor: | Fluorescent protein cyOFP | | Authors: | Sens, A, Ataie, N, Ng, H.L, Lin, M.Z, Chu, J. | | Deposit date: | 2015-05-29 | | Release date: | 2016-06-01 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A Guide to Fluorescent Protein FRET Pairs.
Sensors Basel Sensors, 16, 2016
|
|
6WEM
 
 | | Crimson 0.9 | | Descriptor: | mCrimson 0.9 | | Authors: | Ataie, N, Tran Tang, C, Sens, A, Lin, M.Z, Chu, J, Ng, H.L. | | Deposit date: | 2020-04-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crimson 0.9
To Be Published
|
|
6C68
 
 | | MHC-independent t cell receptor A11 | | Descriptor: | T-cell receptor alpha chain, T-cell receptor beta chain | | Authors: | Lu, J, Van Laethem, F, Saba, I, Chu, J, Bhattacharya, A, Love, N.C, Tikhonova, A, Radaev, S, Sun, X, Ko, A, Arnon, T, Shifrut, E, Friedman, N, Weng, N, Singer, A, Sun, P.D. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of MHC-Independent TCRs and Their Recognition of Native Antigen CD155.
J Immunol., 204, 2020
|
|
7RHB
 
 | |
7RHA
 
 | | A new fluorescent protein darkmRuby at pH 5.0 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a new fluorescent protein darkmRuby at pH 5.0
To Be Published
|
|
7RHC
 
 | | A new fluorescent protein darkmRuby at pH 9.0 | | Descriptor: | 1,2-ETHANEDIOL, darkmRuby | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Long-range Interaction Affects Brightness and pH Stability of a Dark Fluorescent Protein
To Be Published
|
|
7RHD
 
 | | darkmRuby M94T/F96Y mutant at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, darkmRuby M94T/F96Y mutant | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Long-range Interaction Affects Brightness and pH Stability of a Dark Fluorescent Protein
To Be Published
|
|
6M63
 
 | | Crystal structure of a cAMP sensor G-Flamp1. | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Chimera of Cyclic nucleotide-gated potassium channel mll3241 and Yellow fluorescent protein | | Authors: | Zhou, Z, Chen, S, Wang, L, Chu, J. | | Deposit date: | 2020-03-12 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A high-performance genetically encoded fluorescent indicator for in vivo cAMP imaging.
Nat Commun, 13, 2022
|
|
4OQW
 
 | | Crystal structure of mCardinal far-red fluorescent protein | | Descriptor: | Fluorescent protein FP480 | | Authors: | Burg, J.S, Chu, J, Lam, A.J, Lin, M.Z, Garcia, K.C. | | Deposit date: | 2014-02-10 | | Release date: | 2014-03-12 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein.
Nat.Methods, 11, 2014
|
|
5J71
 
 | | Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PS35 | | Descriptor: | 4-({5-[(piperidin-4-yl)amino]-1,3-dihydro-2H-isoindol-2-yl}sulfonyl)benzene-1,3-diol, L(+)-TARTARIC ACID, [Pyruvate dehydrogenase (acetyl-transferring)] kinase isozyme 2, ... | | Authors: | Gui, W.J, Tso, S.C, Chuang, J.L, Wu, C.Y, Qi, X, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2016-04-05 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of Dihydroxyphenyl Sulfonylisoindoline Derivatives as Liver-Targeting Pyruvate Dehydrogenase Kinase Inhibitors.
J. Med. Chem., 60, 2017
|
|
4DZY
 
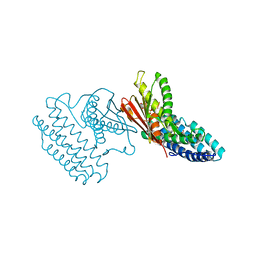 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(S)-2-chloro-3-phenylpropanoic acid complex with ADP | | Descriptor: | (S)-2-chloro-3-phenylpropanoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-03-01 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure-based design and mechanisms of allosteric inhibitors for mitochondrial branched-chain alpha-ketoacid dehydrogenase kinase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1K8M
 
 | | Solution Structure of the Lipoic Acid-Bearing Domain of the E2 component of Human, Mitochondrial Branched-Chain alpha-Ketoacid Dehydrogenase | | Descriptor: | E2 component of Branched-Chain alpha-Ketoacid Dehydrogenase | | Authors: | Chang, C.-F, Chou, H.-T, Chuang, J.L, Chuang, D.T, Huang, T.-h. | | Deposit date: | 2001-10-24 | | Release date: | 2001-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the lipoic acid-bearing domain of human mitochondrial branched-chain alpha-keto acid dehydrogenase complex
J.Biol.Chem., 277, 2002
|
|
1K8O
 
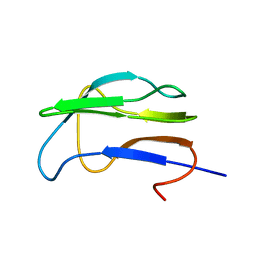 | | Solution Structure of the Lipoic Acid-Bearing Domain of the E2 component of Human, Mitochondrial Branched-Chain alpha-Ketoacid Dehydrogenase | | Descriptor: | E2 component of Branched-Chain alpha-Ketoacid Dehydrogenase | | Authors: | Chang, C.-F, Chou, H.-T, Chuang, J.L, Chuang, D.T, Huang, T.-h. | | Deposit date: | 2001-10-24 | | Release date: | 2001-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the lipoic acid-bearing domain of human mitochondrial branched-chain alpha-keto acid dehydrogenase complex
J.Biol.Chem., 277, 2002
|
|
1U5B
 
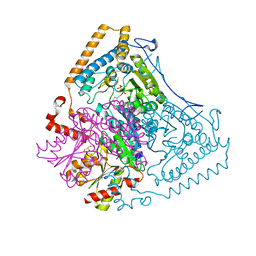 | | Crystal structure of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-oxoisovalerate dehydrogenase alpha subunit, 2-oxoisovalerate dehydrogenase beta subunit, GLYCEROL, ... | | Authors: | Wynn, R.M, Kato, M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-07-27 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Molecular mechanism for regulation of the human mitochondrial branched-chain alpha-ketoacid dehydrogenase complex by phosphorylation
Structure, 12, 2004
|
|
8QFG
 
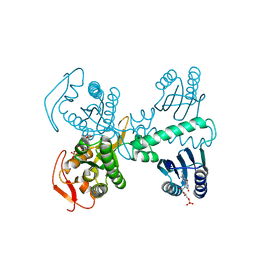 | | Cryogenic crystal structure of the Photoactivated Adenylate Cyclase OaPAC after 5 seconds of blue light illumination | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFE
 
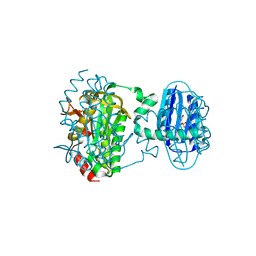 | | Cryogenic crystal structure of the Photoactivated Adenylate Cyclase OaPAC | | Descriptor: | FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, MAGNESIUM ION | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFF
 
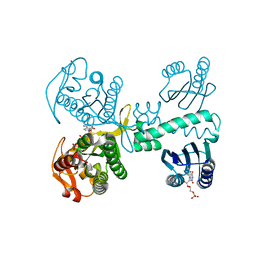 | | Cryogenic crystal structure of the Photoactivated Adenylate Cyclase OaPAC with ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFH
 
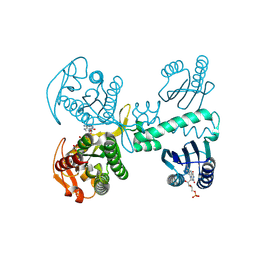 | | Room temperature crystal structure of the Photoactivated Adenylate Cyclase OaPAC with ATP bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFJ
 
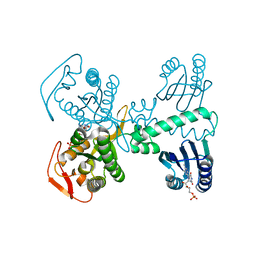 | | Room temperature crystal structure of the Photoactivated Adenylate Cyclase OaPAC after blue light excitation at 2.3 us delay | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Round, A, Sato, T, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
8QFI
 
 | | Room temperature crystal structure of the Photoactivated Adenylate Cyclase OaPAC after blue light excitation at 1.8 us delay | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FLAVIN MONONUCLEOTIDE, Family 3 adenylate cyclase, ... | | Authors: | Chretien, A, Nagel, M.F, Botha, S, de Wijn, R, Brings, L, Doerner, K, Han, H, C.P.Koliyadu, J, Letrun, R, Sato, T, Round, A, Schmidt, C, Secareanu, R, von Stetten, D, Vakili, M, Wrona, A, Bean, R, Mancuso, A, Schulz, J, R.Pearson, A, Kottke, T, Lorenzen, K, Schubert, R. | | Deposit date: | 2023-09-04 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Light-induced Trp in /Met out Switching During BLUF Domain Activation in ATP-bound Photoactivatable Adenylate Cyclase OaPAC.
J.Mol.Biol., 436, 2024
|
|
4D3C
 
 | |
7O3W
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3X
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O40
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3Y
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | Authors: | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
