3GQF
 
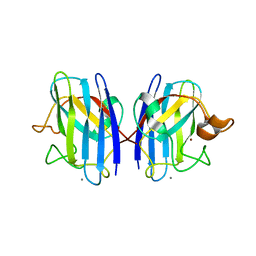 | |
1SZ7
 
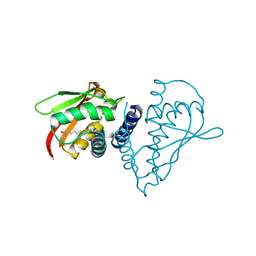 | | Crystal structure of Human Bet3 | | Descriptor: | PALMITIC ACID, Trafficking protein particle complex subunit 3 | | Authors: | Turnbull, A.P, Prinz, B, Holz, C, Behlke, J, Schultchen, J, Delbrueck, H, Niesen, F.H, Lang, C, Heinemann, U. | | Deposit date: | 2004-04-05 | | Release date: | 2005-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of palmitoylated BET3: insights into TRAPP complex assembly and membrane localization
Embo J., 24, 2005
|
|
1TJ1
 
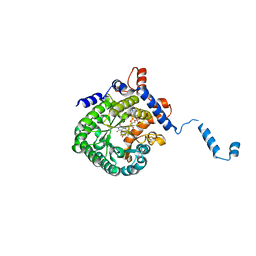 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with L-lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
3GKT
 
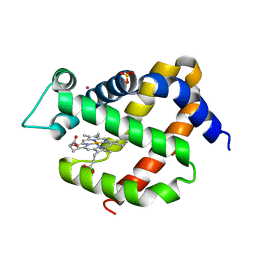 | | Crystal structure of murine neuroglobin under Kr pressure | | Descriptor: | KRYPTON, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Moschetti, T, Mueller, U, Schultze, J, Brunori, M, Vallone, B. | | Deposit date: | 2009-03-11 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of neuroglobin at high Xe and Kr pressure reveals partial conservation of globin internal cavities.
Biophys. J., 97, 2009
|
|
3GLN
 
 | | Carbonmonoxy Ngb under Xenon pressure | | Descriptor: | CARBON MONOXIDE, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Moschetti, T, Mueller, U, Schultze, J, Brunori, M, Vallone, B. | | Deposit date: | 2009-03-12 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The structure of neuroglobin at high Xe and Kr pressure reveals partial conservation of globin internal cavities.
Biophys. J., 97, 2009
|
|
1TJ0
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) co-crystallized with L-lactate | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-02 | | Release date: | 2004-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
1TIW
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with L-Tetrahydro-2-furoic acid | | Descriptor: | Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE, TETRAHYDROFURAN-2-CARBOXYLIC ACID | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-02 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
1TJ2
 
 | | Crystal structure of E. coli PutA proline dehydrogenase domain (residues 86-669) complexed with acetate | | Descriptor: | ACETATE ION, Bifunctional putA protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Tanner, J.J, Zhang, M, White, T.A, Schuermann, J.P, Baban, B.A, Becker, D.F. | | Deposit date: | 2004-06-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors
Biochemistry, 43, 2004
|
|
1IE5
 
 | | NMR STRUCTURE OF THE THIRD IMMUNOGLOBULIN DOMAIN FROM THE NEURAL CELL ADHESION MOLECULE. | | Descriptor: | NEURAL CELL ADHESION MOLECULE | | Authors: | Atkins, A.R, Chung, J, Deechongkit, S, Little, E.B, Edelman, G.M, Wright, P.E, Cunningham, B.A, Dyson, H.J. | | Deposit date: | 2001-04-06 | | Release date: | 2001-08-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third immunoglobulin domain of the neural cell adhesion molecule N-CAM: can solution studies define the mechanism of homophilic binding?
J.Mol.Biol., 311, 2001
|
|
5T2Y
 
 | | Crystal Structure of C. jejuni PglD in complex with 5-methyl-4-(methylamino)-2-phenethylthieno[2,3-d]pyrimidine-6-carboxylic acid | | Descriptor: | 5-methyl-4-(methylamino)-2-(2-phenylethyl)thieno[2,3-d]pyrimidine-6-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | De Schutter, J.W, Imperiali, B. | | Deposit date: | 2016-08-24 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Targeting Bacillosamine Biosynthesis in Bacterial Pathogens: Development of Inhibitors to a Bacterial Amino-Sugar Acetyltransferase from Campylobacter jejuni.
J. Med. Chem., 60, 2017
|
|
