3EXE
 
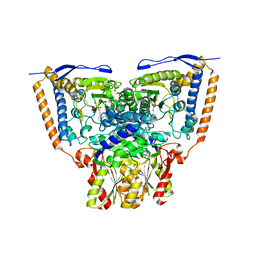 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | GLYCEROL, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
3EXI
 
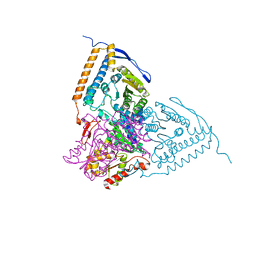 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex with the subunit-binding domain (SBD) of E2p, but SBD cannot be modeled into the electron density | | Descriptor: | CHLORIDE ION, POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
3EXH
 
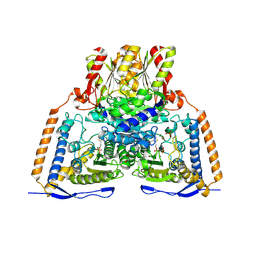 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | GLYCEROL, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
3EXF
 
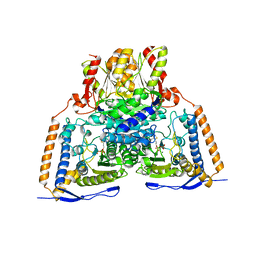 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
3EXG
 
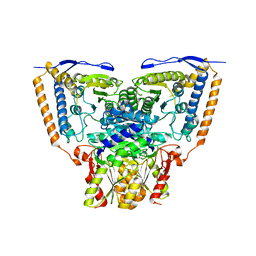 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, somatic form, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
1OLX
 
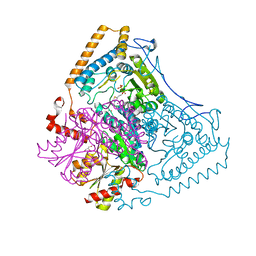 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-18 | | Release date: | 2003-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
6WPU
 
 | |
6X6K
 
 | | Cryo-EM Structure of the Helicobacter pylori dCag3 OMC | | Descriptor: | Cag pathogenicity island protein, Cag pathogenicity island protein (Cag7), Type IV secretion system apparatus protein CagX | | Authors: | Sheedlo, M.J, Chung, J.M, Sawhney, N, Durie, C.L, Cover, T.L, Ohi, M.D, Lacy, D.B. | | Deposit date: | 2020-05-28 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM reveals species-specific components within the Helicobacter pylori Cag type IV secretion system core complex.
Elife, 9, 2020
|
|
6X6J
 
 | | Cryo-EM Structure of CagX and CagY within the Helicobacter pylori PR | | Descriptor: | Cag pathogenicity island protein (Cag7), Cag pathogenicity island protein (Cag8) | | Authors: | Sheedlo, M.J, Chung, J.M, Sawhney, N, Durie, C.L, Cover, T.L, Ohi, M.D, Lacy, D.B. | | Deposit date: | 2020-05-28 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM reveals species-specific components within the Helicobacter pylori Cag type IV secretion system core complex.
Elife, 9, 2020
|
|
6X66
 
 | | Legionella pneumophila dDot T4SS OMC | | Descriptor: | DotC, DotD, Inner membrane lipoprotein YiaD, ... | | Authors: | Durie, C.L, Sheedlo, M.J, Chung, J.M, Byrne, B.G, Su, M, Knight, T, Swanson, M.S, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2020-05-27 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of the Legionella pneumophila Dot/Icm type IV secretion system core complex.
Elife, 9, 2020
|
|
6X6L
 
 | | Cryo-EM Structure of CagX and CagY within the dCag3 Helicobacter pylori PR | | Descriptor: | Cag pathogenicity island protein (Cag7), Cag pathogenicity island protein (Cag8) | | Authors: | Sheedlo, M.J, Chung, J.M, Sawhney, N, Durie, C.L, Cover, T.L, Ohi, M.D, Lacy, D.B. | | Deposit date: | 2020-05-28 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM reveals species-specific components within the Helicobacter pylori Cag type IV secretion system core complex.
Elife, 9, 2020
|
|
6X64
 
 | | Legionella pneumophila Dot T4SS PR | | Descriptor: | Type IV secretion system unknown protein fragment | | Authors: | Durie, C.L, Sheedlo, M.J, Chung, J.M, Byrne, B.G, Su, M, Knight, T, Swanson, M.S, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2020-05-27 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural analysis of the Legionella pneumophila Dot/Icm type IV secretion system core complex.
Elife, 9, 2020
|
|
6X62
 
 | | Legionella pneumophila Dot T4SS OMC | | Descriptor: | DotC, DotD, Inner membrane lipoprotein YiaD, ... | | Authors: | Durie, C.L, Sheedlo, M.J, Chung, J.M, Byrne, B.G, Su, M, Knight, T, Swanson, M.S, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural analysis of the Legionella pneumophila Dot/Icm type IV secretion system core complex.
Elife, 9, 2020
|
|
6X65
 
 | | Legionella pneumophila Dot/Icm T4SS | | Descriptor: | DotC, DotD, Inner membrane lipoprotein YiaD, ... | | Authors: | Durie, C.L, Sheedlo, M.J, Chung, J.M, Byrne, B.G, Su, M, Knight, T, Swanson, M.S, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2020-05-27 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural analysis of the Legionella pneumophila Dot/Icm type IV secretion system core complex.
Elife, 9, 2020
|
|
6X6S
 
 | | Cryo-EM Structure of the Helicobacter pylori OMC | | Descriptor: | Cag pathogenicity island protein (Cag7), Type IV secretion system apparatus protein Cag3, Type IV secretion system apparatus protein CagM, ... | | Authors: | Sheedlo, M.J, Chung, J.M, Sawhney, N, Durie, C.L, Cover, T.L, Ohi, M.D, Lacy, D.B. | | Deposit date: | 2020-05-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM reveals species-specific components within the Helicobacter pylori Cag type IV secretion system core complex.
Elife, 9, 2020
|
|
8QEA
 
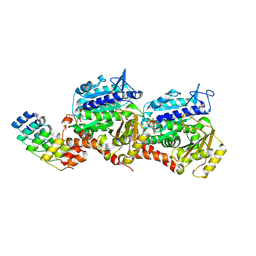 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 96 fs structure | | Descriptor: | Azo-Combretastatin A4 (cis), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-08-31 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QL6
 
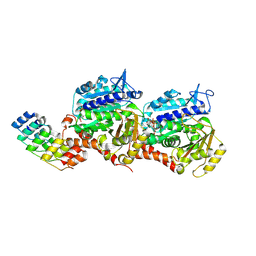 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 25 ps structure | | Descriptor: | Azo-Combretastatin A4 (trans), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QL5
 
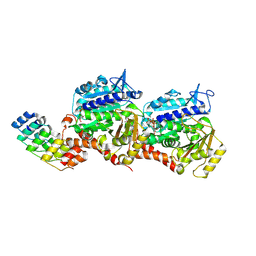 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 1 ps structure | | Descriptor: | Azo-Combretastatin A4 (cis), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QL3
 
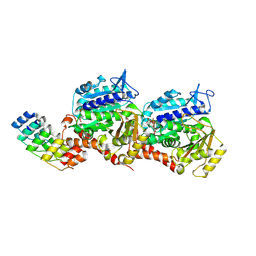 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 233 fs structure | | Descriptor: | Azo-Combretastatin A4 (cis), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QL7
 
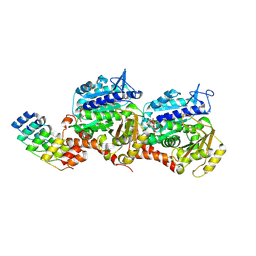 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 35 ps structure | | Descriptor: | Azo-Combretastatin A4 (trans), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QL8
 
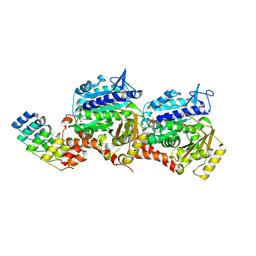 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 125 ps structure | | Descriptor: | Azo-Combretastatin A4 (trans), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QLB
 
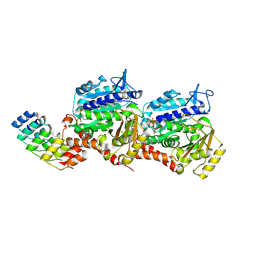 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 100 ns structure | | Descriptor: | Azo-Combretastatin A4 (trans), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QL4
 
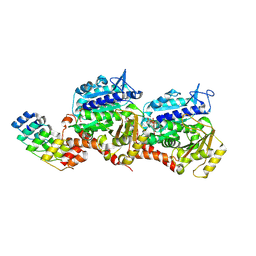 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: 349 fs structure | | Descriptor: | Azo-Combretastatin A4 (cis), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8QL2
 
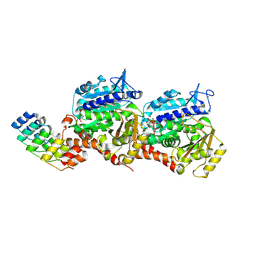 | | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution: dark structure | | Descriptor: | Azo-Combretastatin A4 (cis), CALCIUM ION, Designed Ankyrin Repeat Protein (DARPIN) D1, ... | | Authors: | Weinert, T, Wranik, M, Seidel, H.-P, Church, J, Steinmetz, M.O, Schapiro, I, Standfuss, J. | | Deposit date: | 2023-09-19 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ultrafast structural transitions in an azobenzene photoswitch at near-atomic resolution
To Be Published
|
|
8RJL
 
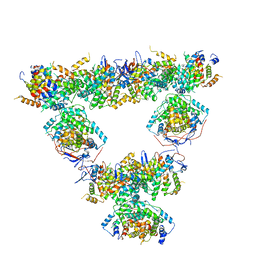 | | Structure of a first order Sierpinski triangle formed by the H369R mutant of the citrate synthase from Synechococcus elongatus | | Descriptor: | Citrate synthase | | Authors: | Lo, Y.K, Bohn, S, Sendker, F.L, Schuller, J.M, Hochberg, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Emergence of fractal geometries in the evolution of a metabolic enzyme.
Nature, 628, 2024
|
|
