1G4B
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
1BOE
 
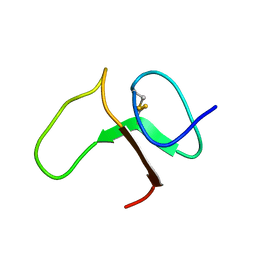 | | STRUCTURE OF THE IGF BINDING DOMAIN OF THE INSULIN-LIKE GROWTH FACTOR-BINDING PROTEIN-5 (IGFBP-5): IMPLICATIONS FOR IGF AND IGF-I RECEPTOR INTERACTIONS | | Descriptor: | PROTEIN (INSULIN-LIKE GROWTH FACTOR-BINDING PROTEIN-5 (IGFBP-5)) | | Authors: | Kalus, W, Zweckstetter, M, Renner, C, Sanchez, Y, Georgescu, J, Grol, M, Demuth, D, Schumacherdony, C, Lang, K, Holak, T.H. | | Deposit date: | 1998-07-30 | | Release date: | 1998-12-16 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the IGF-binding domain of the insulin-like growth factor-binding protein-5 (IGFBP-5): implications for IGF and IGF-I receptor interactions.
EMBO J., 17, 1998
|
|
2Z3B
 
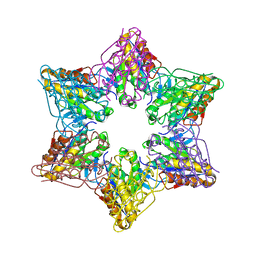 | | Crystal Structure of Bacillus Subtilis CodW, a non-canonical HslV-like peptidase with an impaired catalytic apparatus | | Descriptor: | ATP-dependent protease hslV, SODIUM ION | | Authors: | Rho, S.H, Park, H.H, Kang, G.B, Lim, Y.J, Kang, M.S, Lim, B.K, Seong, I.S, Chung, C.H, Wang, J, Eom, S.H. | | Deposit date: | 2007-06-03 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Bacillus subtilis CodW, a noncanonical HslV-like peptidase with an impaired catalytic apparatus
Proteins, 71, 2007
|
|
4QF1
 
 | | Crystal structure of unliganded CH59UA, the inferred unmutated ancestor of the RV144 anti-HIV antibody lineage producing CH59 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CH59UA Fab fragment of heavy chain, CHLORIDE ION, ... | | Authors: | Wiehe, K, Easterhoff, D, Luo, K, Nicely, N.I, Bradley, T, Jaeger, F.H, Dennison, S.M, Zhang, R, Lloyd, K.E, Stolarchuk, C, Parks, R, Sutherland, L.L, Scearce, R.M, Morris, L, Kaewkungwal, J, Nitayaphan, S, Pitisuttithum, P, Rerks-Ngarm, S, Michael, N, Kim, J, Kelsoe, G, Montefiori, D.C, Tomaras, G, Bonsignori, M, Santra, S, Kepler, T.B, Alam, S.M, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2014-05-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antibody Light-Chain-Restricted Recognition of the Site of Immune Pressure in the RV144 HIV-1 Vaccine Trial Is Phylogenetically Conserved.
Immunity, 41, 2014
|
|
1LM3
 
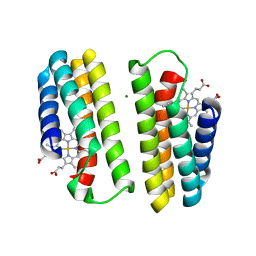 | | A Multi-generation Analysis of Cytochrome b562 Redox Variants: Evolutionary Strategies for Modulating Redox Potential Revealed Using a Library Approach | | Descriptor: | MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, SOLUBLE CYTOCHROME B562 | | Authors: | Springs, S.L, Bass, S.E, Bowman, G, Nodelman, I, Schutt, C.E, McLendon, G.L. | | Deposit date: | 2002-04-30 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A multigeneration analysis of cytochrome b(562) redox variants: evolutionary strategies for modulating redox potential revealed using a library approach.
Biochemistry, 41, 2002
|
|
1CF1
 
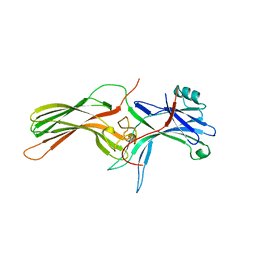 | | ARRESTIN FROM BOVINE ROD OUTER SEGMENTS | | Descriptor: | PROTEIN (ARRESTIN) | | Authors: | Hirsch, J.A, Schubert, C, Gurevich, V.V, Sigler, P.B. | | Deposit date: | 1999-03-23 | | Release date: | 1999-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.8 A crystal structure of visual arrestin: a model for arrestin's regulation.
Cell(Cambridge,Mass.), 97, 1999
|
|
2Z84
 
 | | Insights from crystal and solution structures of mouse UfSP1 | | Descriptor: | Ufm1-specific protease 1 | | Authors: | Ha, B.H, Ahn, H.C, Kang, S.H, Tanaka, K, Chung, C.H, Kim, E.E. | | Deposit date: | 2007-08-30 | | Release date: | 2008-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for Ufm1 processing by UfSP1
J. Biol. Chem., 283, 2008
|
|
1G4A
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | Authors: | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | Deposit date: | 2000-10-26 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
4QYY
 
 | | Discovery of Novel, Dual Mechanism ERK Inhibitors by Affinity Selection Screening of an Inactive Kinase State | | Descriptor: | (3R)-1-{2-[4-(4-acetylphenyl)piperazin-1-yl]-2-oxoethyl}-N-(3-chloro-4-hydroxyphenyl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Deng, Y, Shipps, G.W, Cooper, A, English, J.M, Annis, D.A, Carr, D, Nan, Y, Wang, T, Zhu, Y.H, Chuang, C, Dayananth, P, Hruza, A.W, Xiao, L, Jin, W, Kirschmeier, P, Windsor, W.T, Samatar, A.A. | | Deposit date: | 2014-07-26 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Novel, Dual Mechanism ERK Inhibitors by Affinity Selection Screening of an Inactive Kinase.
J.Med.Chem., 57, 2014
|
|
6J0I
 
 | | Structure of [Co2+-(Chromomycin A3)2]-d(TTGGCGAA)2 complex | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYROXY-7-METHYLANTHRACENE, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Satange, R.B, Chuang, C.Y, Hou, M.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Polymorphic G:G mismatches act as hotspots for inducing right-handed Z DNA by DNA intercalation.
Nucleic Acids Res., 47, 2019
|
|
4G4E
 
 | | Crystal structure of the L88A mutant of HslV from Escherichia coli | | Descriptor: | ATP-dependent protease subunit HslV | | Authors: | Lee, J.W, Park, E, Yoo, H.M, Ha, B.H, An, J.Y, Jeon, Y.J, Seol, J.H, Eom, S.H, Chung, C.H. | | Deposit date: | 2012-07-16 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Structural Alteration in the Pore Motif of the Bacterial 20S Proteasome Homolog HslV Leads to Uncontrolled Protein Degradation
J.Mol.Biol., 425, 2013
|
|
5DUS
 
 | | Crystal structure of MERS-CoV macro domain in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, ORF1a, ... | | Authors: | Cho, C.-C, Lin, M.-H, Chuang, C.-Y, Hsu, C.-H. | | Deposit date: | 2015-09-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Macro Domain from Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Is an Efficient ADP-ribose Binding Module: CRYSTAL STRUCTURE AND BIOCHEMICAL STUDIES
J.Biol.Chem., 291, 2016
|
|
1A0K
 
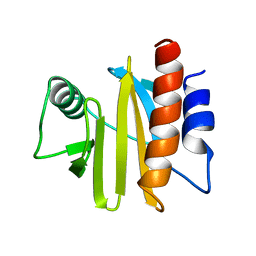 | |
4LR6
 
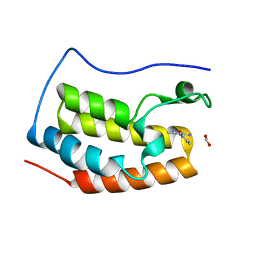 | | Structure of BRD4 bromodomain 1 with a 3-methyl-4-phenylisoxazol-5-amine fragment | | Descriptor: | 3-methyl-4-phenyl-1,2-oxazol-5-amine, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Ravichandran, S, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
2KB6
 
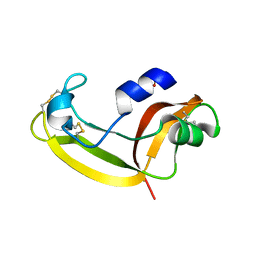 | | Solution structure of onconase C87A/C104A | | Descriptor: | Protein P-30 | | Authors: | Weininger, U, Schulenburg, C, Arnold, U, Ulbrich-Hofmann, R, Balbach, J. | | Deposit date: | 2008-11-21 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Impact of the C-terminal disulfide bond on the folding and stability of onconase.
Chembiochem, 11, 2010
|
|
2Z3A
 
 | | Crystal Structure of Bacillus Subtilis CodW, a non-canonical HslV-like peptidase with an impaired catalytic apparatus | | Descriptor: | ATP-dependent protease hslV | | Authors: | Rho, S.H, Park, H.H, Kang, G.B, Lim, Y.J, Kang, M.S, Lim, B.K, Seong, I.S, Chung, C.H, Wang, J, Eom, S.H. | | Deposit date: | 2007-06-03 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Bacillus subtilis CodW, a noncanonical HslV-like peptidase with an impaired catalytic apparatus
Proteins, 71, 2007
|
|
3FD7
 
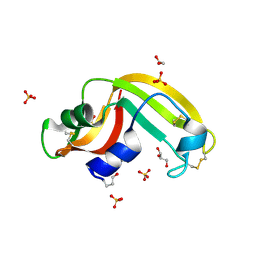 | | Crystal structure of Onconase C87A/C104A-ONC | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein P-30, ... | | Authors: | Neumann, P, Schulenburg, C, Arnold, U, Ulbrich-Hofmann, R, Stubbs, M.T. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Impact of the C-terminal disulfide bond on the folding and stability of onconase.
Chembiochem, 11, 2010
|
|
4LRG
 
 | | Structure of BRD4 bromodomain 1 with a dimethyl thiophene isoxazole azepine carboxamide | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-[1,2]oxazolo[5,4-c]thieno[2,3-e]azepin-6-yl]acetamide, Bromodomain-containing protein 4 | | Authors: | Ravichandran, S, Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
1SOJ
 
 | | CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 3B IN COMPLEX WITH IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, MAGNESIUM ION, cGMP-inhibited 3',5'-cyclic phosphodiesterase B | | Authors: | Scapin, G, Patel, S.B, Chung, C, Varnerin, J.P, Edmondson, S.D, Mastracchio, A, Parmee, E.R, Becker, J.W, Singh, S.B, Van Der Ploeg, L.H, Tota, M.R. | | Deposit date: | 2004-03-15 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Human Phosphodiesterase 3B: Atomic Basis for Substrate and Inhibitor Specificity
Biochemistry, 43, 2004
|
|
1SO2
 
 | | CATALYTIC DOMAIN OF HUMAN PHOSPHODIESTERASE 3B In COMPLEX WITH A DIHYDROPYRIDAZINE INHIBITOR | | Descriptor: | 1-DEOXY-1-[(2-HYDROXYETHYL)(NONANOYL)AMINO]HEXITOL, 6-(4-{[2-(3-IODOBENZYL)-3-OXOCYCLOHEX-1-EN-1-YL]AMINO}PHENYL)-5-METHYL-4,5-DIHYDROPYRIDAZIN-3(2H)-ONE, MAGNESIUM ION, ... | | Authors: | Scapin, G, Patel, S.B, Chung, C, Varnerin, J.P, Edmondson, S.D, Mastracchio, A, Parmee, E.R, Becker, J.W, Singh, S.B, Van Der Ploeg, L.H, Tota, M.R. | | Deposit date: | 2004-03-12 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Human Phosphodiesterase 3B: Atomic Basis for Substrate and Inhibitor Specificity
Biochemistry, 43, 2004
|
|
1HT2
 
 | | Nucleotide-Dependent Conformational Changes in a Protease-Associated ATPase HslU | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK LOCUS HSLU, HEAT SHOCK LOCUS HSLV | | Authors: | Wang, J, Song, J.J, Seong, I.S, Franklin, M.C, Kamtekar, S, Eom, S.H, Chung, C.H. | | Deposit date: | 2000-12-27 | | Release date: | 2001-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU.
Structure, 9, 2001
|
|
2R2X
 
 | | Ricin A-chain (recombinant) complex with Urea | | Descriptor: | Ricin A chain, SULFATE ION, UREA | | Authors: | Carra, J.H, McHugh, C.A, Mulligan, S, Machiesky, L.M, Soares, A.S, Millard, C.B. | | Deposit date: | 2007-08-28 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based identification of determinants of conformational and spectroscopic change at the ricin active site.
Bmc Struct.Biol., 7, 2007
|
|
1T4F
 
 | | Structure of human MDM2 in complex with an optimized p53 peptide | | Descriptor: | SULFATE ION, Ubiquitin-protein ligase E3 Mdm2, optimized p53 peptide | | Authors: | Grasberger, B.L, Schubert, C, Koblish, H.K, Carver, T.E, Franks, C.F, Zhao, S.Y, Lu, T, LaFrance, L.V, Parks, D.J. | | Deposit date: | 2004-04-29 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells
J.Med.Chem., 48, 2005
|
|
1T4E
 
 | | Structure of Human MDM2 in complex with a Benzodiazepine Inhibitor | | Descriptor: | (4-CHLOROPHENYL)[3-(4-CHLOROPHENYL)-7-IODO-2,5-DIOXO-1,2,3,5-TETRAHYDRO-4H-1,4-BENZODIAZEPIN-4-YL]ACETIC ACID, Ubiquitin-protein ligase E3 Mdm2 | | Authors: | Grasberger, B.L, Schubert, C, Koblish, H.K, Carver, T.E, Franks, C.F, Zhao, S.Y, Lu, T, LaFrance, L.V, Parks, D.J. | | Deposit date: | 2004-04-29 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells
J.Med.Chem., 48, 2005
|
|
4IR4
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with 1-[7-(morpholin-4-yl)-1-(pyridin-2-yl)indolizin-3-yl]ethanone (GSK2834113A) | | Descriptor: | 1,2-ETHANEDIOL, 1-[7-(morpholin-4-yl)-1-(pyridin-2-yl)indolizin-3-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Chung, C.W, Drewry, D, Chen, P, Filippakopoulos, P, Fedorov, O, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B.
J.Med.Chem., 59, 2016
|
|
