4IGV
 
 | | Crystal structure of kirola (Act d 11) | | Descriptor: | CHLORIDE ION, Kirola, UNKNOWN LIGAND | | Authors: | Chruszcz, M, Ciardiello, M.A, Giangrieco, I, Osinski, T, Minor, W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins.
Mol.Immunol., 56, 2013
|
|
4IH0
 
 | | Crystal structure of kirola (Act d 11) from crystal soaked with serotonin | | Descriptor: | CHLORIDE ION, Kirola, MAGNESIUM ION, ... | | Authors: | Chruszcz, M, Ciardiello, M.A, Giangrieco, I, Osinski, T, Minor, W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins.
Mol.Immunol., 56, 2013
|
|
4IH2
 
 | | Crystal structure of kirola (Act d 11) from crystal soaked with 2-aminopurine | | Descriptor: | CHLORIDE ION, Kirola, UNKNOWN LIGAND | | Authors: | Chruszcz, M, Ciardiello, M.A, Giangrieco, I, Osinski, T, Minor, W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and bioinformatic analysis of the kiwifruit allergen Act d 11, a member of the family of ripening-related proteins.
Mol.Immunol., 56, 2013
|
|
3RVT
 
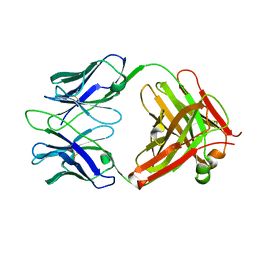 | | Structure of 4C1 Fab in P212121 space group | | Descriptor: | Fab fragment of 4C1 antibody - heavy chain, Fab fragment of 4C1 antibody - light chain | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2011-05-06 | | Release date: | 2012-01-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular determinants for antibody binding on group 1 house dust mite allergens.
J.Biol.Chem., 287, 2012
|
|
3RVU
 
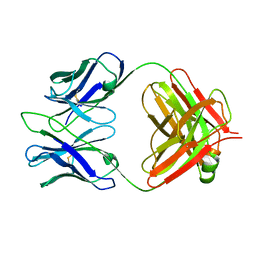 | | Structure of 4C1 Fab in C2221 space group | | Descriptor: | 4C1 Fab - heavy chain, 4C1 Fab - light chain | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2011-05-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular determinants for antibody binding on group 1 house dust mite allergens.
J.Biol.Chem., 287, 2012
|
|
3S7I
 
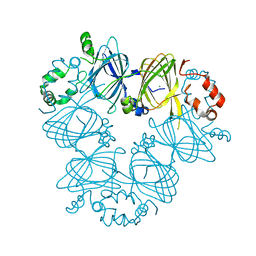 | | Crystal structure of Ara h 1 | | Descriptor: | Allergen Ara h 1, clone P41B, CHLORIDE ION | | Authors: | Chruszcz, M, Maleki, S.J, Solberg, R, Minor, W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Immunologic Characterization of Ara h 1, a Major Peanut Allergen.
J.Biol.Chem., 286, 2011
|
|
3S7E
 
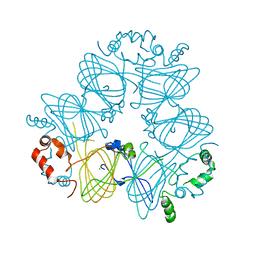 | | Crystal structure of Ara h 1 | | Descriptor: | Allergen Ara h 1, clone P41B, CHLORIDE ION | | Authors: | Chruszcz, M, Maleki, S.J, Solberg, R, Minor, W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural and Immunologic Characterization of Ara h 1, a Major Peanut Allergen.
J.Biol.Chem., 286, 2011
|
|
3PZW
 
 | | Soybean lipoxygenase-1 - re-refinement | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FE (II) ION, ... | | Authors: | Chruszcz, M, Minor, W. | | Deposit date: | 2010-12-14 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Determination of protein structures - a series of fortunate events.
Biophys.J., 95, 2008
|
|
7KSB
 
 | | Crystal structure on Act c 10.0101 | | Descriptor: | Non-specific lipid-transfer protein 1, SULFATE ION | | Authors: | Pote, S, O'Malley, A, Gawlicka-Chruszcz, A, Giangrieco, I, Ciardiello, M.A, Chruszcz, M. | | Deposit date: | 2020-11-21 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Characterization of Act c 10.0101 and Pun g 1.0101-Allergens from the Non-Specific Lipid Transfer Protein Family.
Molecules, 26, 2021
|
|
7KSC
 
 | | Crystal structure of Pun g 1.0101 | | Descriptor: | Non-specific lipid-transfer protein, SULFATE ION | | Authors: | Pote, S, O'Malley, A, Gawlicka-Chruszcz, A, Tuppo, L, Ciardiello, M.A, Chruszcz, M. | | Deposit date: | 2020-11-21 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Characterization of Act c 10.0101 and Pun g 1.0101-Allergens from the Non-Specific Lipid Transfer Protein Family.
Molecules, 26, 2021
|
|
8GKN
 
 | | Crystal structure of TuUGT202A2 (Tetur22g00270) in complex with S-naringenin | | Descriptor: | NARINGENIN, UDP-glycosyltransferase 202A2, URIDINE-5'-DIPHOSPHATE | | Authors: | Arriaza, R.H, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2023-03-20 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of TuUGT202A2 (Tetur22g00270) in complex with S-naringenin
To Be Published
|
|
4N7C
 
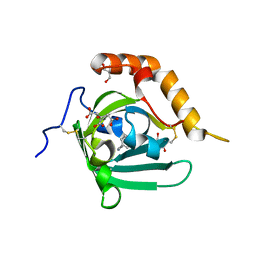 | | Structural re-examination of native Bla g 4 | | Descriptor: | 4-(2-aminoethyl)phenol, Bla g 4 allergen variant 1, CITRIC ACID, ... | | Authors: | Offermann, L.R, Chan, S.L, Osinski, T, Tan, Y.W, Chew, F.T, Sivaraman, J, Mok, Y.K, Minor, W, Chruszcz, M. | | Deposit date: | 2013-10-15 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The major cockroach allergen Bla g 4 binds tyramine and octopamine.
Mol.Immunol., 60, 2014
|
|
3KZL
 
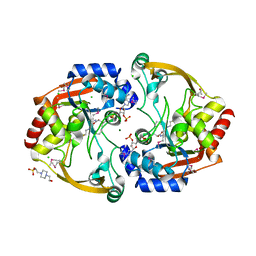 | | Crystal structure of BA2930 mutant (H183G) in complex with AcCoA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Porebski, P.J, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-08 | | Release date: | 2009-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
3K3S
 
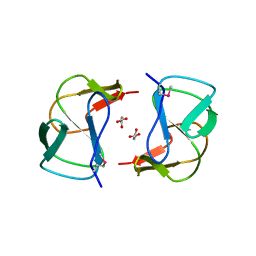 | | Crystal structure of altronate hydrolase (fragment 1-84) from Shigella Flexneri. | | Descriptor: | ACETATE ION, Altronate hydrolase, CHLORIDE ION, ... | | Authors: | Hou, J, Chruszcz, M, Xu, X, Le, B, Zimmerman, M.D, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-04 | | Release date: | 2009-10-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of altronate hydrolase (fragment 1-84) from Shigella Flexneri.
To be Published
|
|
7LVY
 
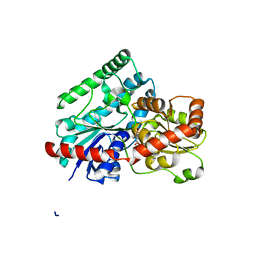 | | Crystal Structure of Tetur04g02350 | | Descriptor: | UDP-glycosyltransferase 203A2, URIDINE-5'-DIPHOSPHATE, beta-D-glucopyranose | | Authors: | Danehsian, L, Kluza, A, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2021-02-26 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Tetur04g02350
To Be Published
|
|
2PZ9
 
 | | Crystal structure of putative transcriptional regulator SCO4942 from Streptomyces coelicolor | | Descriptor: | Putative regulatory protein, SULFATE ION | | Authors: | Filippova, E.V, Chruszcz, M, Xu, X, Zheng, H, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-17 | | Release date: | 2007-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In situ proteolysis for protein crystallization and structure determination.
Nat.Methods, 4, 2007
|
|
2Q24
 
 | | Crystal structure of TetR transcriptional regulator SCO0520 from Streptomyces coelicolor | | Descriptor: | ACETATE ION, CHLORIDE ION, Putative tetR family transcriptional regulator | | Authors: | Cymborowski, M, Chruszcz, M, Koclega, K.D, Filippova, E.V, Xu, X, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-25 | | Release date: | 2007-07-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a putative transcriptional regulator SCO0520 from Streptomyces coelicolor A3(2) reveals an unusual dimer among TetR family proteins.
J.Struct.Funct.Genom., 12, 2011
|
|
6XO2
 
 | | Structural Characterization of Beta Cyanoalanine Synthase from Tetranychus Urticae (two-spotted spider mite) | | Descriptor: | ACETATE ION, Beta-cyanoalanine synthase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Daneshian, L, Schlachter, C, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional characterization of beta-cyanoalanine synthase from Tetranychus urticae.
Insect Biochem.Mol.Biol., 142, 2022
|
|
1UXO
 
 | | The crystal structure of the ydeN gene product from B. subtilis | | Descriptor: | Putative hydrolase YdeN | | Authors: | Janda, I.K, Devedjiev, Y, Cooper, D.R, Chruszcz, M, Derewenda, U, Gabrys, A, Minor, W, Joachimiak, A, Derewenda, Z.S, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-27 | | Release date: | 2004-05-27 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Harvesting the high-hanging fruit: the structure of the YdeN gene product from Bacillus subtilis at 1.8 angstroms resolution.
Acta Crystallogr. D Biol. Crystallogr., 60, 2004
|
|
6MBX
 
 | |
4U12
 
 | | Crystal structure of protein HP0242 from Helicobacter pylori at 1.94 A resolution: a knotted homodimer | | Descriptor: | Uncharacterized protein HP0242 | | Authors: | Grabowski, M, Shabalin, I.G, Chruszcz, M, Skarina, T, Onopriyenko, O, Guthrie, J, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-14 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of protein HP0242 from Helicobacter pylori at 1.94 A resolution: a knotted homodimer
to be published
|
|
6OY4
 
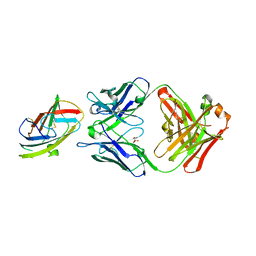 | | Crystal structure of complex between recombinant Der p 2.0103 and Fab fragment of 7A1 | | Descriptor: | Der p 2 variant 3, Fab fragment of IgG, HEAVY CHAIN, ... | | Authors: | Kapingidza, A.B, Offermann, L.R, Glesner, J, Wunschmann, S, Vailes, L.D, Chapman, M.D.C, Pomes, A, Chruszcz, M. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Human IgE Antibody Binding Site on Der p 2 for the Design of a Recombinant Allergen for Immunotherapy.
J Immunol., 203, 2019
|
|
8STX
 
 | |
6PMU
 
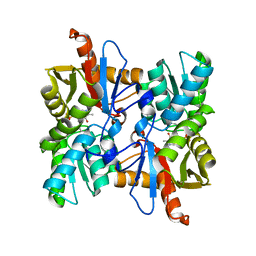 | | Structural Characterization of Beta Cyanoalanine Synthase from Tetranychus Urticae | | Descriptor: | Beta-cyanoalanine synthase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Daneshian, L, Schlachter, C, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of Beta Cyanoalanine Synthase from Tetranychus Urticae
To Be Published
|
|
6PNT
 
 | |
