4KOV
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cefuroxime | | Descriptor: | (6R,7R)-3-[(carbamoyloxy)methyl]-7-{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Otwinowski, Z, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4MAP
 
 | | Crystal structure of Ara h 8 purified with heating | | Descriptor: | Ara h 8 allergen, SODIUM ION | | Authors: | Offermann, L.R, Hurlburt, B.K, Majorek, K.A, McBride, J.K, Maleki, S.J, Chruszcz, M. | | Deposit date: | 2013-08-16 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Function of the Peanut Panallergen Ara h 8.
J.Biol.Chem., 288, 2013
|
|
4KOU
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with Cefixime | | Descriptor: | (6R,7R)-7-({(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-[(carboxymethoxy)imino]acetyl}amino)-3-ethenyl-8-oxo-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid, 1,2-ETHANEDIOL, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Otwinowski, Z, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, Functional, and Inhibition Studies of a Gcn5-related N-Acetyltransferase (GNAT) Superfamily Protein PA4794: A NEW C-TERMINAL LYSINE PROTEIN ACETYLTRANSFERASE FROM PSEUDOMONAS AERUGINOSA.
J.Biol.Chem., 288, 2013
|
|
4MA6
 
 | | Crystal structure of Ara h 8 with Epicatechin bound | | Descriptor: | (2R,3R)-2-(3,4-dihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol, Ara h 8 allergen, SODIUM ION | | Authors: | Offermann, L.R, Hurlburt, B.K, Majorek, K.A, McBride, J.K, Maleki, S.J, Chruszcz, M. | | Deposit date: | 2013-08-15 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of the Peanut Panallergen Ara h 8.
J.Biol.Chem., 288, 2013
|
|
4M3S
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Majorek, K.A, Chruszcz, M, Xu, X, Cymborowski, M, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-08-06 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Double trouble-Buffer selection and His-tag presence may be responsible for nonreproducibility of biomedical experiments.
Protein Sci., 23, 2014
|
|
3TYS
 
 | | Crystal structure of transcriptional regulator VanUg, Form II | | Descriptor: | Predicted transcriptional regulator | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Depardieu, F, Courvalin, P, Shabalin, I, Chruszcz, M, Minor, W, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.121 Å) | | Cite: | Crystal structure of transcriptional regulator VanUg, Form II
TO BE PUBLISHED
|
|
3P7J
 
 | | Drosophila HP1a chromo shadow domain | | Descriptor: | GLYCEROL, Heterochromatin protein 1 | | Authors: | Kim, D, Chruszcz, M, Minor, W, Khorasanizadeh, S. | | Deposit date: | 2010-10-12 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The HP1a Disordered C Terminus and Chromo Shadow Domain Cooperate to Select Target Peptide Partners.
Chembiochem, 12, 2011
|
|
3QTB
 
 | | Structure of the universal stress protein from Archaeoglobus fulgidus in complex with dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ACETATE ION, Uncharacterized protein | | Authors: | Tkaczuk, K.L, Shumilin, I.A, Chruszcz, M, Cymborowski, M, Xu, X, Di Leo, R, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
4JRM
 
 | | Crystal structure of beta-ketoacyl-ACP synthase II (FabF) from Vibrio Cholerae (space group P212121) at 1.75 Angstrom | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, ACETATE ION, GLYCEROL | | Authors: | Hou, J, Chruszcz, M, Shabalin, I.G, Zheng, H, Cooper, D.R, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-21 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of beta-ketoacyl-ACP synthase II (FabF) from Vibrio cholerae (space group P43) at 2.2 Angstrom
To be Published
|
|
3UDO
 
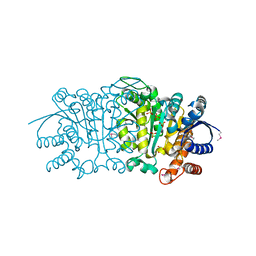 | | Crystal structure of putative isopropylamlate dehydrogenase from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, 3-isopropylmalate dehydrogenase, SULFATE ION | | Authors: | Tkaczuk, K.L, Chruszcz, M, Blus, B.J, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-28 | | Release date: | 2011-11-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of putative isopropylamlate dehydrogenase from Campylobacter jejuni
To be Published
|
|
4M9B
 
 | | Crystal structure of Apo Ara h 8 | | Descriptor: | Ara h 8 allergen, SODIUM ION | | Authors: | Offermann, L.R, Hurlburt, B.K, Majorek, K.A, McBride, J.K, Maleki, S.J, Chruszcz, M. | | Deposit date: | 2013-08-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Function of the Peanut Panallergen Ara h 8.
J.Biol.Chem., 288, 2013
|
|
4M9W
 
 | | Crystal Structure of Ara h 8 with MES bound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ara h 8 allergen, SODIUM ION | | Authors: | Offermann, L.R, Hurlburt, B.K, Majorek, K.A, McBride, J.K, Maleki, S.J, Chruszcz, M. | | Deposit date: | 2013-08-15 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Function of the Peanut Panallergen Ara h 8.
J.Biol.Chem., 288, 2013
|
|
3TNJ
 
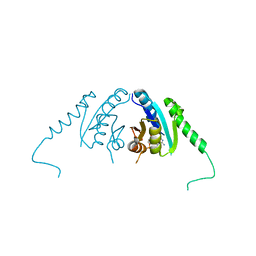 | | Crystal structure of universal stress protein from Nitrosomonas europaea with AMP bound | | Descriptor: | ADENOSINE MONOPHOSPHATE, Universal stress protein (Usp) | | Authors: | Tkaczuk, K.L, Chruszcz, M, Shumilin, I.A, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-01 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
3QTD
 
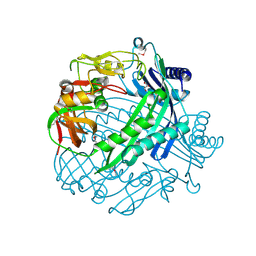 | | Crystal structure of putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa PAO1 | | Descriptor: | GLYCEROL, PmbA protein | | Authors: | Tkaczuk, K.L, Chruszcz, M, Evdokimova, E, Liu, F, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of putative modulator of gyrase (PmbA) from Pseudomonas aeruginosa PAO1
To be Published
|
|
3QSL
 
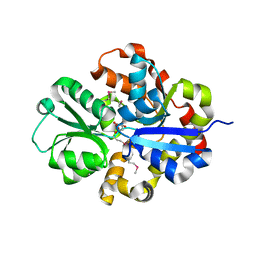 | | Structure of CAE31940 from Bordetella bronchiseptica RB50 | | Descriptor: | CITRIC ACID, Putative exported protein | | Authors: | Bajor, J, Kagan, O, Chruszcz, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-21 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of pyrimidine/thiamin biosynthesis precursor-like domain-containing protein CAE31940 from proteobacterium Bordetella bronchiseptica RB50, and evolutionary insight into the NMT1/THI5 family.
J Struct Funct Genomics, 15, 2014
|
|
3OWC
 
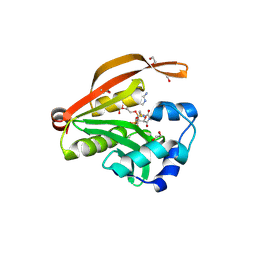 | | Crystal structure of GNAT superfamily protein PA2578 from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, Probable acetyltransferase | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-09-17 | | Release date: | 2010-11-03 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of GNAT superfamily protein PA2578 from Pseudomonas aeruginosa
To be Published
|
|
5DQF
 
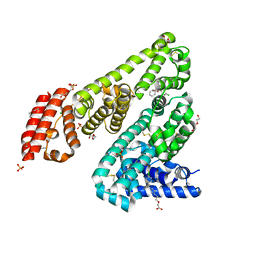 | | Horse Serum Albumin (ESA) in complex with Cetirizine | | Descriptor: | (2-{4-[(R)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}ethoxy)acetic acid, (2-{4-[(S)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}ethoxy)acetic acid, CHLORIDE ION, ... | | Authors: | Handing, K.B, Shabalin, I.G, Majorek, K.A, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-09-14 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of equine serum albumin in complex with cetirizine reveals a novel drug binding site.
Mol.Immunol., 71, 2016
|
|
2DG2
 
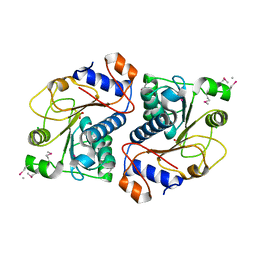 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein | | Descriptor: | Apolipoprotein A-I binding protein, CHLORIDE ION, SULFATE ION | | Authors: | Shumilin, I.A, Jha, K.N, Zheng, H, Chruszcz, M, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2006-03-08 | | Release date: | 2007-03-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Biochemical and structural characterization of apolipoprotein A-I binding protein, a novel phosphoprotein with a potential role in sperm capacitation.
Endocrinology, 149, 2008
|
|
3DM8
 
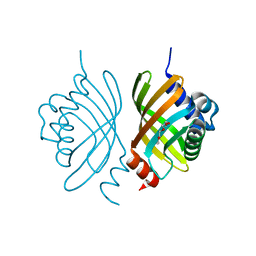 | | Crystal Structure of Putative Isomerase from Rhodopseudomonas palustris | | Descriptor: | DODECYL NONA ETHYLENE GLYCOL ETHER, uncharacterized protein RPA4348 | | Authors: | Cymborowski, M, Chruszcz, M, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Putative Isomerase from Rhodopseudomonas palustris
To be Published
|
|
3LNL
 
 | | Crystal structure of Staphylococcus aureus protein SA1388 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, UPF0135 protein SA1388, ZINC ION | | Authors: | Singh, K.S, Chruszcz, M, Zhang, X, Minor, W, Zhang, H. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Conserved Hypothetical Protein Sa1388 from S. aureus Reveals a Capped Hexameric Toroid with Two Pii Domain Lids and a Dinuclear Metal Center.
Bmc Struct.Biol., 6, 2006
|
|
3O4F
 
 | | Crystal Structure of Spermidine Synthase from E. coli | | Descriptor: | SULFATE ION, Spermidine synthase | | Authors: | Zhou, X, Tkaczuk, K.L, Chruszcz, M, Chua, T.K, Minor, W, Sivaraman, J. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of Escherichia coli spermidine synthase SpeE reveals a unique substrate-binding pocket
J.Struct.Biol., 169, 2010
|
|
2ID3
 
 | | Crystal structure of transcriptional regulator SCO5951 from Streptomyces coelicolor A3(2) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative transcriptional regulator | | Authors: | Grabowski, M, Chruszcz, M, Koclega, K.D, Cymborowski, M, Gu, J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-14 | | Release date: | 2006-10-17 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: |
|
|
2PAQ
 
 | | Crystal structure of the 5'-deoxynucleotidase YfbR | | Descriptor: | 5'-deoxynucleotidase YfbR | | Authors: | Zimmerman, M.D, Chruszcz, M, Cymborowski, M, Kudritska, M, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-27 | | Release date: | 2007-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: the 5'-deoxyribonucleotidase YfbR from Escherichia coli.
J.Mol.Biol., 378, 2008
|
|
2O8N
 
 | | Crystal Structure of Mouse Apolipoprotein A-I Binding Protein | | Descriptor: | ApoA-I binding protein, CHLORIDE ION, SULFATE ION | | Authors: | Shumilin, I.A, Jha, K.N, Zheng, H, Chruszcz, M, Cymborowski, M, Herr, J.C, Minor, W. | | Deposit date: | 2006-12-12 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and Structural Characterization of Apolipoprotein A-I Binding Protein, a Novel Phosphoprotein with a Potential Role in Sperm Capacitation.
Endocrinology, 149, 2008
|
|
3IRC
 
 | | Crystal structure analysis of dengue-1 envelope protein domain III | | Descriptor: | ENVELOPE PROTEIN, SULFATE ION | | Authors: | Nelson, C.A, Kim, T, Warren, J.T, Chruszcz, M, Minor, W, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure Analysis of the Dengue-1 Envelope Protein Domain III
To be Published
|
|
