4PGJ
 
 | |
8V3K
 
 | | F95S-F198S epi-Isozizaene Synthase: complex with 3 Mg2+, inorganic pyrophosphate, and benzyl triethyl ammonium cation | | Descriptor: | DIPHOSPHATE, Epi-isozizaene synthase, GLYCEROL, ... | | Authors: | Christianson, D.W, Eaton, S.A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Based Engineering of a Sesquiterpene Cyclase to Generate an Alcohol Product: Conversion of epi -Isozizaene Synthase into alpha-Bisabolol Synthase.
Biochemistry, 63, 2024
|
|
6V0K
 
 | |
6BMB
 
 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase (T381G mutant) in complex with tartrate and shikimate | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic, ... | | Authors: | Christendat, D, Peek, J. | | Deposit date: | 2017-11-14 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.077 Å) | | Cite: | Structural and biochemical approaches uncover multiple evolutionary trajectories of plant quinate dehydrogenases.
Plant J., 2018
|
|
6BMQ
 
 | | Crystal structure of Arabidopsis Dehydroquinate dehydratase-shikimate dehydrogenase (T381G mutant) in complex with tartrate and shikimate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase, chloroplastic, ... | | Authors: | Christendat, D, Peek, J. | | Deposit date: | 2017-11-15 | | Release date: | 2018-09-26 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.077 Å) | | Cite: | Structural and biochemical approaches uncover multiple evolutionary trajectories of plant quinate dehydrogenases.
Plant J., 2018
|
|
3PWZ
 
 | | Crystal structure of an Ael1 enzyme from Pseudomonas putida | | Descriptor: | Shikimate dehydrogenase 3 | | Authors: | Christendat, D, Peek, J. | | Deposit date: | 2010-12-09 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Structural and mechanistic analysis of a novel class of shikimate dehydrogenases: evidence for a conserved catalytic mechanism in the shikimate dehydrogenase family.
Biochemistry, 50, 2011
|
|
1L1S
 
 | | Structure of Protein of Unknown Function MTH1491 from Methanobacterium thermoautotrophicum | | Descriptor: | hypothetical protein MTH1491 | | Authors: | Christendat, D, Saridakis, V, Kim, Y, Kumar, P.A, Xu, X, Semesi, A, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-19 | | Release date: | 2002-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of hypothetical protein MTH1491 from Methanobacterium thermoautotrophicum.
Protein Sci., 11, 2002
|
|
1EIJ
 
 | | NMR ENSEMBLE OF METHANOBACTERIUM THERMOAUTOTROPHICUM PROTEIN 1615 | | Descriptor: | HYPOTHETICAL PROTEIN MTH1615 | | Authors: | Christendat, D, Booth, V, Gernstein, M, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-25 | | Release date: | 2000-11-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
2O7S
 
 | |
2O7Q
 
 | |
2NNG
 
 | | Structure of inhibitor binding to Carbonic Anhydrase II | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONAMIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-05-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
2NNS
 
 | |
2NNO
 
 | | Structure of inhibitor binding to Carbonic Anhydrase II | | Descriptor: | 3-[4-(AMINOSULFONYL)PHENYL]PROPANOIC ACID, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-05-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
2NNV
 
 | | Structure of inhibitor binding to Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase 2, ETHYL 3-[4-(AMINOSULFONYL)PHENYL]PROPANOATE, GLYCEROL, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-24 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
2NN7
 
 | | Structure of inhibitor binding to Carbonic Anhydrase I | | Descriptor: | Carbonic anhydrase 1, DIMETHYL SULFOXIDE, ETHYL 3-[4-(AMINOSULFONYL)PHENYL]PROPANOATE, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-23 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II.
J.Am.Chem.Soc., 129, 2007
|
|
2NN1
 
 | | Structure of inhibitor binding to Carbonic Anhydrase I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[4-(AMINOSULFONYL)PHENYL]PROPANOIC ACID, Carbonic anhydrase 1, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II
J.Am.Chem.Soc., 129, 2007
|
|
1KJN
 
 | |
1EP0
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-24 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
2NMX
 
 | | Structure of inhibitor binding to Carbonic Anhydrase I | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Carbonic anhydrase 1, N-{2-[4-(AMINOSULFONYL)PHENYL]ETHYL}ACETAMIDE, ... | | Authors: | Christianson, D.W, Jude, K.M. | | Deposit date: | 2006-10-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Analysis of Charge Discrimination in the Binding of Inhibitors to Human Carbonic Anhydrases I and II
J.Am.Chem.Soc., 129, 2007
|
|
1EPZ
 
 | | CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOASE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM WITH BOUND LIGAND. | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C, Edwards, A.M. | | Deposit date: | 2000-03-30 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
1EJE
 
 | | CRYSTAL STRUCTURE OF AN FMN-BINDING PROTEIN | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, NICKEL (II) ION, ... | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Arrowsmith, C, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
8CWJ
 
 | |
8CWK
 
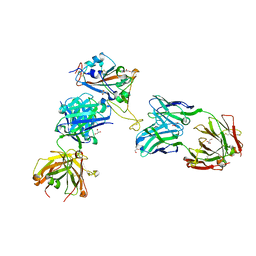 | |
8CWI
 
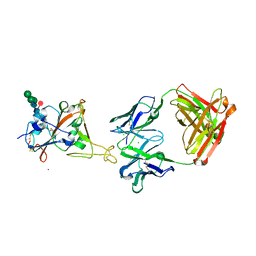 | | Fab arm of antibody 10G4 bound to CoV-2 receptor binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Heavy chain of Fab arm of antibody 10G4, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.873 Å) | | Cite: | Neutralization of CoV-2 omicron lineages by affinity-matured class 5 antibodies
To Be Published
|
|
6MH2
 
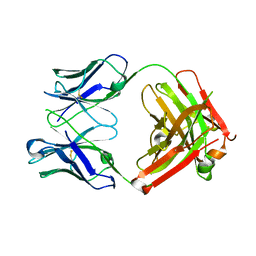 | | Structure of Herceptin Fab without antigen | | Descriptor: | Herceptin Fab arm heavy chain, Herceptin Fab arm light chain | | Authors: | Luthra, A, Langley, D.B, Christie, M, Christ, D. | | Deposit date: | 2018-09-17 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human Antibody Bispecifics through Phage Display Selection.
Biochemistry, 58, 2019
|
|
