2OHY
 
 | |
2IWY
 
 | | Human mitochondrial beta-ketoacyl ACP synthase | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE, AMMONIUM ION | | Authors: | Christensen, C.E, Kragelund, B.B, von Wettstein-Knowles, P, Henriksen, A. | | Deposit date: | 2006-07-05 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of the Human Beta-Ketoacyl [Acp] Synthase from the Mitochondrial Type II Fatty Acid Synthase.
Protein Sci., 16, 2007
|
|
2IX4
 
 | | Arabidopsis thaliana mitochondrial beta-ketoacyl ACP synthase hexanoic acid complex | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE, HEXANOIC ACID, POTASSIUM ION | | Authors: | Christensen, C.E, Kragelund, B.B, von Wettstein-Knowles, P, Henriksen, A. | | Deposit date: | 2006-07-06 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Human Beta-Ketoacyl [Acp] Synthase from the Mitochondrial Type II Fatty Acid Synthase.
Protein Sci., 16, 2007
|
|
2YG2
 
 | | Structure of apolioprotein M in complex with Sphingosine 1-Phosphate | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, APOLIPOPROTEIN M, CITRATE ANION | | Authors: | Christoffersen, C, Obinata, H, Gowda, S, Galvani, S, Ahnstrom, J, Sevvana, M, Egerer-Sieber, C, Muller, Y.A, Hla, T, Nielsen, L, Dahlback, B. | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Endothelium-Protective Sphingosine-1-Phosphate Provided by Hdl-Associated Apolipoprotein M.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2IWZ
 
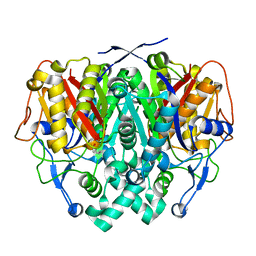 | | Human mitochondrial beta-ketoacyl ACP synthase complexed with hexanoic acid | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE, AMMONIUM ION, HEXANOIC ACID | | Authors: | Christensen, C.E, Kragelund, B.B, von Wettstein-Knowles, P, Henriksen, A. | | Deposit date: | 2006-07-05 | | Release date: | 2007-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the Human Beta-Ketoacyl [Acp] Synthase from the Mitochondrial Type II Fatty Acid Synthase.
Protein Sci., 16, 2007
|
|
2QVE
 
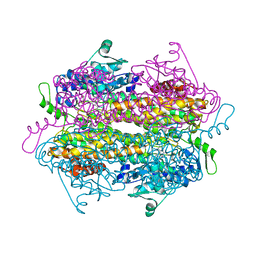 | |
6YM5
 
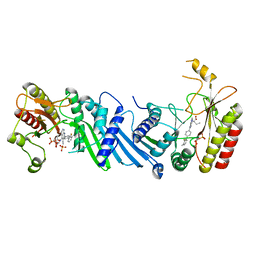 | | Crystal structure of BAY-091 with PIP4K2A | | Descriptor: | (2~{R})-2-[[3-cyano-2-[4-(2-fluoranyl-3-methyl-phenyl)phenyl]-1,7-naphthyridin-4-yl]amino]butanoic acid, PHOSPHATE ION, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
6YM3
 
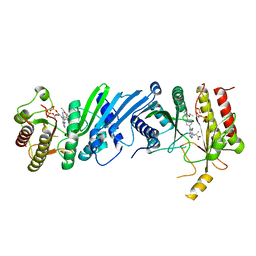 | | Crystal structure of Compound 1 with PIP4K2A | | Descriptor: | (2~{R})-2-[[3-cyano-2-[4-(2-ethoxyphenyl)phenyl]-5,8-dihydro-1,7-naphthyridin-4-yl]amino]propanoic acid, PHOSPHATE ION, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
6YM4
 
 | | Crystal structure of BAY-297 with PIP4K2A | | Descriptor: | (2~{R})-2-[[2-[4-(3-chloranyl-2-fluoranyl-phenyl)phenyl]-3-cyano-1,7-naphthyridin-4-yl]amino]butanamide, GLYCEROL, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha | | Authors: | Holton, S.J, Wortmann, L, Braeuer, N, Irlbacher, H, Weiske, J, Lechner, C, Meier, R, Puetter, V, Christ, C, ter Laak, T, Lienau, P, Lesche, R, Nicke, B, Bauser, M, Haegebarth, A, von Nussbaum, F, Mumberg, D, Lemos, C. | | Deposit date: | 2020-04-07 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Characterization of the Potent and Highly Selective 1,7-Naphthyridine-Based Inhibitors BAY-091 and BAY-297 of the Kinase PIP4K2A.
J.Med.Chem., 64, 2021
|
|
5N49
 
 | | BRPF2 in complex with Compound 7 | | Descriptor: | 2-(1,3,6-trimethyl-2-oxidanylidene-benzimidazol-5-yl)benzo[de]isoquinoline-1,3-dione, Bromodomain-containing protein 1 | | Authors: | Bouche, L, Christ, C.D, Siegel, S, Fernandez-Montalvan, A.E, Holton, S.J, Fedorov, O, ter Laak, A, Sugawara, T, Stoeckigt, D, Tallant, C, Bennett, J, Monteiro, O, Saez, L.D, Siejka, P, Meier, J, Puetter, V, Weiske, J, Mueller, S, Huber, K.V.M, Hartung, I.V, Haendler, B. | | Deposit date: | 2017-02-10 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains.
J. Med. Chem., 60, 2017
|
|
5ARG
 
 | | SMYD2 in complex with SGC probe BAY-598 | | Descriptor: | GLYCEROL, N-LYSINE METHYLTRANSFERASE SMYD2, N-[1-(N'-CYANO-N-[3-(DIFLUOROMETHOXY)PHENYL]CARBAMIMIDOYL)-3-(3,4-DICHLOROPHENYL)-4,5-DIHYDRO-1H-PYRAZOL-4-YL]-N-ETHYL-2-HYDROXYACETAMIDE, ... | | Authors: | Hillig, R.C, Badock, V, Barak, N, Stellfeld, T, Eggert, E, ter Laak, A, Weiske, J, Christ, C.D, Koehr, S, Stoeckigt, D, Mowat, J, Mueller, T, Fernandez-Montalvan, A.E, Hartung, I.V, Stresemann, C, Brumby, T, Weinmann, H. | | Deposit date: | 2015-09-24 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery and Characterization of a Highly Potent and Selective Aminopyrazoline-Based in Vivo Probe (Bay-598) for the Protein Lysine Methyltransferase Smyd2.
J.Med.Chem., 59, 2016
|
|
5ARF
 
 | | SMYD2 in complex with small molecule inhibitor compound-2 | | Descriptor: | N-LYSINE METHYLTRANSFERASE SMYD2, N-[3-(4-CHLOROPHENYL)-1-{N'-CYANO-N-[3-(DIFLUOROMETHOXY)PHENYL]CARBAMIMIDOYL}-4,5-DIHYDRO-1H- PYRAZOL-4-YL]-N-ETHYL-2-HYDROXYACETAMIDE, S-ADENOSYLMETHIONINE, ... | | Authors: | Hillig, R.C, Badock, V, Barak, N, Stellfeld, T, Eggert, E, ter Laak, A, Weiske, J, Christ, C.D, Koehr, S, Stoeckigt, D, Mowat, J, Mueller, T, Fernandez-Montalvan, A.E, Hartung, I.V, Stresemann, C, Brumby, T, Weinmann, H. | | Deposit date: | 2015-09-24 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery and Characterization of a Highly Potent and Selective Aminopyrazoline-Based in Vivo Probe (Bay-598) for the Protein Lysine Methyltransferase Smyd2.
J.Med.Chem., 59, 2016
|
|
5KJ9
 
 | |
2BGQ
 
 | | apo aldose reductase from barley | | Descriptor: | ALDOSE REDUCTASE, SULFATE ION | | Authors: | Olsen, J.G, Pedersen, L, Christensen, C.L, Olsen, O, Henriksen, A. | | Deposit date: | 2005-01-04 | | Release date: | 2006-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Barley Aldose Reductase: Structure, Cofactor Binding, and Substrate Recognition in the Aldo/Keto Reductase 4C Family.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
2BGS
 
 | | HOLO ALDOSE REDUCTASE FROM BARLEY | | Descriptor: | ALDOSE REDUCTASE, BICARBONATE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Olsen, J.G, Pedersen, L, Christensen, C.L, Olsen, O, Henriksen, A. | | Deposit date: | 2005-01-05 | | Release date: | 2006-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Barley Aldose Reductase: Structure, Cofactor Binding, and Substrate Recognition in the Aldo/Keto Reductase 4C Family.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
2RJS
 
 | | SgTAM bound to substrate mimic | | Descriptor: | (3R)-3-amino-2,2-difluoro-3-(4-methoxyphenyl)propanoic acid, Tyrosine aminomutase | | Authors: | Montavon, T.J, Christianson, C.V, Bruner, S.D. | | Deposit date: | 2007-10-15 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and characterization of mechanism-based inhibitors for the tyrosine aminomutase SgTAM.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2RJR
 
 | | Substrate mimic bound to SgTAM | | Descriptor: | (2S,3S)-3-(4-fluorophenyl)-2,3-dihydroxypropanoic acid, Tyrosine aminomutase | | Authors: | Montavon, T.J, Christianson, C.V, Bruner, S.D. | | Deposit date: | 2007-10-15 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and characterization of mechanism-based inhibitors for the tyrosine aminomutase SgTAM.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2Y0J
 
 | | Triazoloquinazolines as a novel class of phosphodiesterase 10A (PDE10A) inhibitors, part 2, Lead-optimisation. | | Descriptor: | 5-(1H-BENZIMIDAZOL-2-YLMETHYLSULFANYL)-2-METHYL-[1,2,4]TRIAZOLO[1,5-C]QUINAZOLINE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE 10A, ... | | Authors: | Kehler, J, Ritzen, A, Langgard, M, Petersen, S.L, Christoffersen, C.T, Nielsen, J, Kilburn, J.P. | | Deposit date: | 2010-12-03 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Triazoloquinazolines as a Novel Class of Phosphodiesterase 10A (Pde10A) Inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7THI
 
 | | Human Bisphosphoglycerate Mutase complexed with 2-phosphoglycolate | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Bisphosphoglycerate mutase | | Authors: | Clark, K.L, Kulathila, R, Wright, K, Isome, Y, Sage, D, Yang, Y, Christodoulou, C. | | Deposit date: | 2022-01-11 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Human Bisphosphoglycerate Mutase complexed with 2-phosphoglycolate
To Be Published
|
|
2WU9
 
 | | Crystal structure of peroxisomal KAT2 from Arabidopsis thaliana | | Descriptor: | 1,2-ETHANEDIOL, 3-KETOACYL-COA THIOLASE 2, PEROXISOMAL | | Authors: | Pye, V.E, Christensen, C.E, Dyer, J.H, Arent, S, Henriksen, A. | | Deposit date: | 2009-10-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Peroxisomal Plant 3-Ketoacyl-Coa Thiolases Structure and Activity are Regulated by a Sensitive Redox Switch
J.Biol.Chem., 285, 2010
|
|
2WUA
 
 | | Structure of the peroxisomal 3-ketoacyl-CoA thiolase from Sunflower | | Descriptor: | ACETOACETYL COA THIOLASE | | Authors: | Pye, V.E, Christensen, C.E, Dyer, J.H, Arent, S, Henriksen, A. | | Deposit date: | 2009-10-01 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peroxisomal Plant 3-Ketoacyl-Coa Thiolases Structure and Activity are Regulated by a Sensitive Redox Switch
J.Biol.Chem., 285, 2010
|
|
2VDG
 
 | | Barley Aldose Reductase 1 complex with butanol | | Descriptor: | 1-BUTANOL, ALDOSE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Olsen, J.G, Pedersen, L, Christensen, C.L, Olsen, O, Henriksen, A. | | Deposit date: | 2007-10-08 | | Release date: | 2008-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Barley Aldose Reductase: Structure, Cofactor Binding, and Substrate Recognition in the Aldo/Keto Reductase 4C Family.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
3QSB
 
 | | Structure of E. coli polIIIbeta with (Z)-5-(1-((4'-Fluorobiphenyl-4-yl)methoxyimino)butyl)-2,2-dimethyl-4,6-dioxocyclohexanecarbonitrile | | Descriptor: | (1R,5R)-5-{(1Z)-N-[(4'-fluorobiphenyl-4-yl)methoxy]butanimidoyl}-2,2-dimethyl-4,6-dioxocyclohexanecarbonitrile, DNA polymerase III subunit beta | | Authors: | Wijffels, G, Johnson, W.M, Oakley, A.J, Turner, K, Epa, V.C, Briscoe, S.J, Polley, M, Liepa, A.J, Hofmann, A, Buchardt, J, Christensen, C, Prosselkov, P, Dalrymple, B.P, Alewood, P.F, Jennings, P.A, Dixon, N.E, Winkler, D.A. | | Deposit date: | 2011-02-20 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding inhibitors of the bacterial sliding clamp by design
J.Med.Chem., 54, 2011
|
|
4IUW
 
 | | Crystal structure of PEPO from Lactobacillus rhamnosis HN001 (DR20) | | Descriptor: | CARBONATE ION, CITRIC ACID, Neutral endopeptidase, ... | | Authors: | Anderson, B.F, Knapp, K.M, Holland, R, Norris, G.E, Christensson, C, Bratt, H, Collins, L.J, Coolbear, T, Lubbers, M.W, Toole, P.W.O, Reid, J.R, Jameson, G.B. | | Deposit date: | 2013-01-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of PEPO from Lactobacillus rhamnosis HN001 (DR20)
TO BE PUBLISHED
|
|
2WTB
 
 | | Arabidopsis thaliana multifuctional protein, MFP2 | | Descriptor: | FATTY ACID MULTIFUNCTIONAL PROTEIN (ATMFP2) | | Authors: | Arent, S, Pye, V.E, Christensen, C.E, Norgaard, A, Henriksen, A. | | Deposit date: | 2009-09-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Multi-Functional Protein in Peroxisomal Beta-Oxidation. Structure and Substrate Specificity of the Arabidopsis Thaliana Protein, Mfp2
J.Biol.Chem., 285, 2010
|
|
