4F3H
 
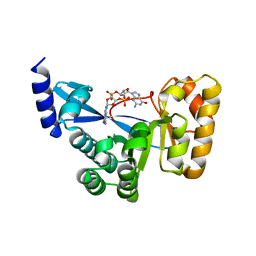 | | The structural of FimXEAL-c-di-GMP from Xanthomonas campestris | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative uncharacterized protein | | Authors: | Chin, K.-H, Liao, Y.-T, Chou, S.-H. | | Deposit date: | 2012-05-09 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural polymorphism of c-di-GMP bound to an EAL domain and in complex with a type II PilZ-domain protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F48
 
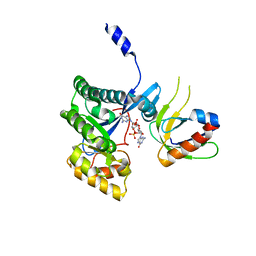 | | The X-ray structural of FimXEAL-c-di-GMP-PilZ complexes from Xanthomonas campestris | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative uncharacterized protein, Type IV fimbriae assembly protein | | Authors: | Chin, K.-H, Liao, Y.-T, Chou, S.-H. | | Deposit date: | 2012-05-10 | | Release date: | 2012-11-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural polymorphism of c-di-GMP bound to an EAL domain and in complex with a type II PilZ-domain protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1P0U
 
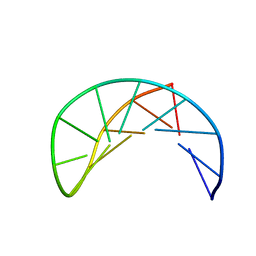 | | Sheared G/C Base Pair | | Descriptor: | 5'-D(*GP*CP*AP*TP*CP*GP*AP*CP*GP*AP*TP*GP*C)-3' | | Authors: | Chin, K.H, Chou, S.H. | | Deposit date: | 2003-04-11 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Sheared-type G(anti).C(syn) Base-pair: A Unique d(GXC) Loop Closure Motif.
J.Mol.Biol., 329, 2003
|
|
8UXW
 
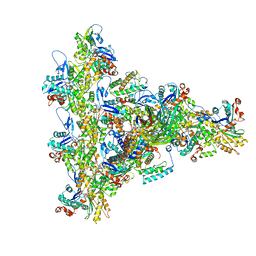 | | Arp2/3 branch junction complex, ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | Deposit date: | 2023-11-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
8UXX
 
 | | Arp2/3 branch junction complex, BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | Deposit date: | 2023-11-11 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
8UZ0
 
 | | Straight actin filament from Arp2/3 branch junction sample (ADP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | Deposit date: | 2023-11-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
8UZ1
 
 | | Straight actin filament from Arp2/3 branch junction sample (ADP-BeFx) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Chavali, S.S, Chou, S.Z, Sindelar, C.V. | | Deposit date: | 2023-11-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures reveal how phosphate release from Arp3 weakens actin filament branches formed by Arp2/3 complex.
Nat Commun, 15, 2024
|
|
4YS2
 
 | | RCK domain with CDA | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Na+/H+ antiporter-like protein | | Authors: | Chin, K.H, Chou, S.H. | | Deposit date: | 2015-03-16 | | Release date: | 2016-04-27 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | RCK domain with CDA
To Be Published
|
|
3IWZ
 
 | | The c-di-GMP Responsive Global Regulator CLP Links Cell-Cell Signaling to Virulence Gene Expression in Xanthomonas campestris | | Descriptor: | Catabolite activation-like protein | | Authors: | Chin, K.H, Tu, Z.L, Tseng, Y.H, Dow, J.M, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2009-09-03 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris.
J.Mol.Biol., 396, 2010
|
|
3PH1
 
 | |
1L3M
 
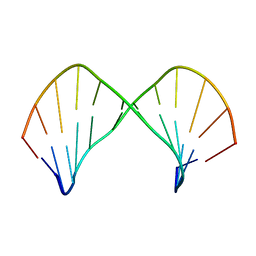 | | The Solution Structure of [d(CGC)r(amamam)d(TTTGCG)]2 | | Descriptor: | 5'-D(*CP*GP*C)-R(P*(A39)P*(A39)P*(A39))-D(P*TP*TP*TP*GP*CP*G)-3' | | Authors: | Tsao, Y.P, Wang, L.Y, Hsu, S.T, Jain, M.L, Chou, S.H, Huang, W.C, Cheng, J.W. | | Deposit date: | 2002-02-28 | | Release date: | 2002-04-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of [d(CGC)r(amamam)d(TTTGCG)]2.
J.Biomol.NMR, 21, 2001
|
|
3DR2
 
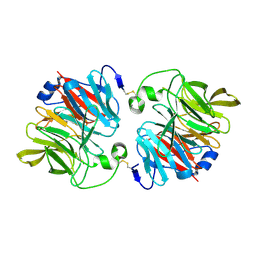 | | Structural and Functional Analyses of XC5397 from Xanthomonas campestris: A Gluconolactonase Important in Glucose Secondary Metabolic Pathways | | Descriptor: | CALCIUM ION, Exported gluconolactonase | | Authors: | Chen, C.-N, Chin, K.-H, Wang, A.H.-J, Chou, S.H. | | Deposit date: | 2008-07-10 | | Release date: | 2008-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The First Crystal Structure of Gluconolactonase Important in the Glucose Secondary Metabolic Pathways
J.Mol.Biol., 384, 2008
|
|
5H5O
 
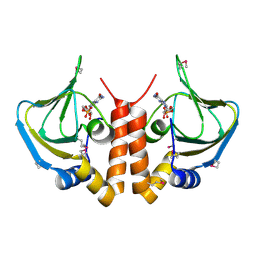 | |
2E12
 
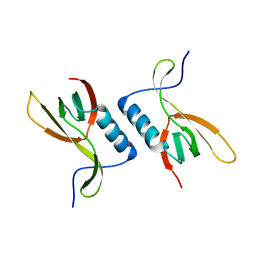 | | The crystal structure of XC5848 from Xanthomonas campestris adopting a novel variant of Sm-like motif | | Descriptor: | Hypothetical protein XCC3642 | | Authors: | Chin, K.-H, Ruan, S.-K, Wang, A.H.-J, Chou, S.-H. | | Deposit date: | 2006-10-17 | | Release date: | 2007-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | XC5848, an ORFan protein from Xanthomonas campestris, adopts a novel variant of Sm-like motif
Proteins, 68, 2007
|
|
1D68
 
 | |
4KBY
 
 | | mSTING/c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Stimulator of interferon genes protein | | Authors: | Chin, K.H, Su, Y.C, Tu, J.L, Chou, S.H. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Novel c-di-GMP recognition modes of the mouse innate immune adaptor protein STING
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4KC0
 
 | | mSTING | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Chin, K.H, Su, Y.C, Tu, J.L, Chou, S.H. | | Deposit date: | 2013-04-24 | | Release date: | 2013-05-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel c-di-GMP recognition modes of the mouse innate immune adaptor protein STING
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2E11
 
 | | The Crystal Structure of XC1258 from Xanthomonas campestris: A CN-hydrolase Superfamily Protein with an Arsenic Adduct in the Active Site | | Descriptor: | CACODYLATE ION, Hydrolase | | Authors: | Chin, K.-H, Tsai, Y.-D, Chan, N.-L, Huang, K.-F, Wang, A.H.-J, Chou, S.-H. | | Deposit date: | 2006-10-17 | | Release date: | 2007-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The crystal structure of XC1258 from Xanthomonas campestris: A putative procaryotic Nit protein with an arsenic adduct in the active site
Proteins, 69, 2007
|
|
2ZE3
 
 | |
4KT3
 
 | | Structure of a type VI secretion system effector-immunity complex from Pseudomonas protegens | | Descriptor: | Putative lipoprotein, Uncharacterized protein | | Authors: | Whitney, J.C, Chou, S, Gardiner, T.E, Mougous, J.D. | | Deposit date: | 2013-05-19 | | Release date: | 2013-07-31 | | Last modified: | 2013-10-09 | | Method: | X-RAY DIFFRACTION (1.4362 Å) | | Cite: | Identification, Structure, and Function of a Novel Type VI Secretion Peptidoglycan Glycoside Hydrolase Effector-Immunity Pair.
J.Biol.Chem., 288, 2013
|
|
3EK5
 
 | | Unique GTP-binding Pocket and Allostery of UMP Kinase from a Gram-Negative Phytopathogen Bacterium | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Uridylate kinase | | Authors: | Tu, J.-L, Chin, K.-H, Wang, A.H.-J, Chou, S.-H. | | Deposit date: | 2008-09-18 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Unique GTP-Binding Pocket and Allostery of Uridylate Kinase from a Gram-Negative Phytopathogenic Bacterium
J.Mol.Biol., 385, 2009
|
|
3EK6
 
 | |
3ROF
 
 | | Crystal Structure of the S. aureus Protein Tyrosine Phosphatase PtpA | | Descriptor: | Expression tag cleaved from protein-tyrosine-phosphatase ptpA, Low molecular weight protein-tyrosine-phosphatase ptpA, PHOSPHATE ION | | Authors: | Grundner, C, Chou, S, Engel, K. | | Deposit date: | 2011-04-25 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure and substrate recognition of the Staphylococcus aureus protein tyrosine phosphatase PtpA.
J.Mol.Biol., 413, 2011
|
|
3C12
 
 | |
1XS3
 
 | | Solution Structure Analysis of the XC975 protein | | Descriptor: | hypothetical protein XC975 | | Authors: | Chin, K.-H, Lin, F.-Y, Hu, Y.-C, Sze, K.-H, Lyu, P.-C, Chou, S.-H. | | Deposit date: | 2004-10-18 | | Release date: | 2005-03-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Letter to the Editor: NMR structure note - Solution structure of a bacterial BolA-like protein XC975 from a plant pathogen Xanthomonas campestris pv. campestris
J.Biomol.Nmr, 31, 2005
|
|
