6NHW
 
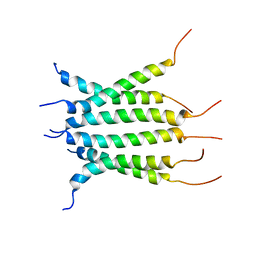 | | Structure of the transmembrane domain of the Death Receptor 5 - Dimer of Trimer | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Fu, Q, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
6NHY
 
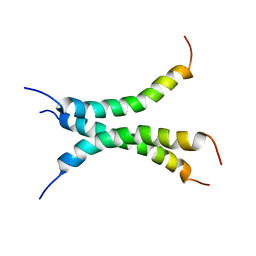 | | Structure of the transmembrane domain of the Death Receptor 5 mutant (G217Y) - Trimer Only | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H, Liu, Z. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
1F71
 
 | |
1F70
 
 | |
1J7O
 
 | |
1J7P
 
 | |
2RLF
 
 | |
2BID
 
 | | HUMAN PRO-APOPTOTIC PROTEIN BID | | Descriptor: | PROTEIN (BID) | | Authors: | Chou, J.J, Li, H, Salvesen, G.S, Yuan, J, Wagner, G. | | Deposit date: | 1999-01-27 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BID, an intracellular amplifier of apoptotic signaling.
Cell(Cambridge,Mass.), 96, 1999
|
|
3CRD
 
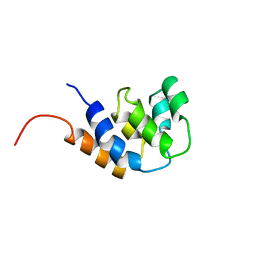 | | NMR STRUCTURE OF THE RAIDD CARD DOMAIN, 15 STRUCTURES | | Descriptor: | RAIDD | | Authors: | Chou, J.J, Matsuo, H, Duan, H, Wagner, G. | | Deposit date: | 1998-07-24 | | Release date: | 1999-02-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment.
Cell(Cambridge,Mass.), 94, 1998
|
|
2HAC
 
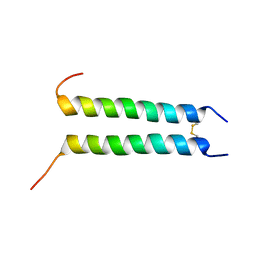 | | Structure of Zeta-Zeta Transmembrane Dimer | | Descriptor: | T-cell surface glycoprotein CD3 zeta chain | | Authors: | Chou, J.J, Wucherpfennig, K.W, Schnell, J.R, Call, M.E. | | Deposit date: | 2006-06-12 | | Release date: | 2006-10-31 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
2N4X
 
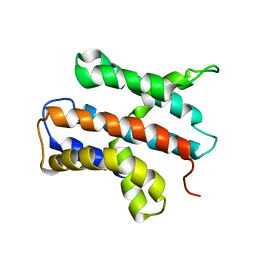 | |
4MMM
 
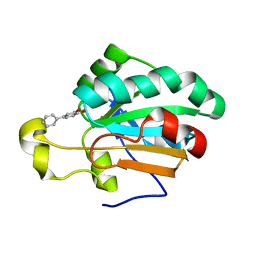 | | Human Pdrx5 complex with a ligand BP7 | | Descriptor: | 1,1'-BIPHENYL-3,4-DIOL, Peroxiredoxin-5, mitochondrial | | Authors: | Guichou, J.F. | | Deposit date: | 2013-09-09 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Comparing Binding Modes of Analogous Fragments Using NMR in Fragment-Based Drug Design: Application to PRDX5
Plos One, 9, 2014
|
|
8PON
 
 | | TEAD2 in complex with an inhibitor | | Descriptor: | 4-fluoranyl-2-[(3-phenylmethoxyphenyl)amino]benzoic acid, MYRISTIC ACID, Transcriptional enhancer factor TEF-4 | | Authors: | Guichou, J.F. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
8POJ
 
 | | TEAD2 in complex with an inhibitor | | Descriptor: | 4-fluoranyl-2-[(3-phenylphenyl)amino]benzoic acid, MYRISTIC ACID, Transcriptional enhancer factor TEF-4 | | Authors: | Guichou, J.F. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
8POM
 
 | | TEAD2 in complex with an inhibitor | | Descriptor: | 2-[[3-(2-phenylethoxy)phenyl]amino]pyridine-3-carboxylic acid, GLYCEROL, Transcriptional enhancer factor TEF-4 | | Authors: | Guichou, J.F. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
8PUX
 
 | | TEAD2 with a covalent inhibitor | | Descriptor: | MYRISTIC ACID, Transcriptional enhancer factor TEF-4, ~{N}-[2-[(3-pentoxyphenyl)amino]phenyl]propanamide | | Authors: | Guichou, J.F, Gelin, M, Allemand, F. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Development of HC-258, a Covalent Acrylamide TEAD Inhibitor That Reduces Gene Expression and Cell Migration.
Acs Med.Chem.Lett., 14, 2023
|
|
8PUY
 
 | | TEAD2 with a covalent inhibitor | | Descriptor: | MYRISTIC ACID, Transcriptional enhancer factor TEF-4, ~{N}-[3-[(3-pentoxyphenyl)amino]phenyl]propanamide | | Authors: | Guichou, J.F, Gelin, M, Allemand, F. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of HC-258, a Covalent Acrylamide TEAD Inhibitor That Reduces Gene Expression and Cell Migration.
Acs Med.Chem.Lett., 14, 2023
|
|
8P29
 
 | | TEAD2 in complex with an inhibitor | | Descriptor: | 5-methyl-2-[(3-phenylmethoxyphenyl)amino]benzoic acid, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Guichou, J.F, Gelin, M, Allemand, F. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
4OOW
 
 | | HCV NS5B polymerase with a fragment of quercetagetin | | Descriptor: | CATECHOL, RNA-directed RNA polymerase | | Authors: | Guichou, J.F, Ahmed-Belkacem, A, Rozenn, B, Nazim, N, Hernandez, E, Pallier, C, Pawlotsky, J.M. | | Deposit date: | 2014-02-04 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Inhibition of RNA binding to hepatitis C virus RNA-dependent RNA polymerase: a new mechanism for antiviral intervention.
Nucleic Acids Res., 42, 2014
|
|
4K7I
 
 | | HUMAN PEROXIREDOXIN 5 with a fragment | | Descriptor: | CATECHOL, DIMETHYL SULFOXIDE, Peroxiredoxin-5, ... | | Authors: | Guichou, J.F. | | Deposit date: | 2013-04-17 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Comparing Binding Modes of Analogous Fragments Using NMR in Fragment-Based Drug Design: Application to PRDX5
Plos One, 9, 2014
|
|
4K7O
 
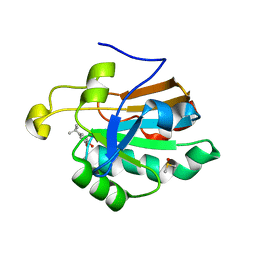 | | HUMAN PEROXIREDOXIN 5 with a fragment | | Descriptor: | 4-tert-butylbenzene-1,2-diol, DIMETHYL SULFOXIDE, Peroxiredoxin-5, ... | | Authors: | Guichou, J.F. | | Deposit date: | 2013-04-17 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Comparing Binding Modes of Analogous Fragments Using NMR in Fragment-Based Drug Design: Application to PRDX5
Plos One, 9, 2014
|
|
4K7N
 
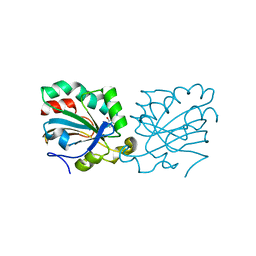 | | HUMAN PEROXIREDOXIN 5 with a fragment | | Descriptor: | 4-METHYLCATECHOL, Peroxiredoxin-5, mitochondrial | | Authors: | Guichou, J.F. | | Deposit date: | 2013-04-17 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparing Binding Modes of Analogous Fragments Using NMR in Fragment-Based Drug Design: Application to PRDX5
Plos One, 9, 2014
|
|
6UJV
 
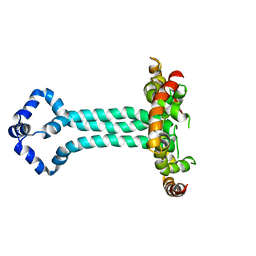 | | Model of the HIV-1 gp41 membrane-proximal external region, transmembrane domain and cytoplasmic tail (LLP2) | | Descriptor: | Envelope glycoprotein GP41 | | Authors: | Piai, A, Fu, Q, Cai, Y, Ghantous, F, Xiao, T, Shaik, M.M, Peng, H, Rits-Volloch, S, Liu, Z, Chen, W, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of transmembrane coupling of the HIV-1 envelope glycoprotein.
Nat Commun, 11, 2020
|
|
7LC8
 
 | | SARS-CoV-2 spike Protein TM domain | | Descriptor: | Spike protein S2' | | Authors: | Fu, Q, Chou, J.J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Trimeric Hydrophobic Zipper Mediates the Intramembrane Assembly of SARS-CoV-2 Spike.
J.Am.Chem.Soc., 143, 2021
|
|
7LOI
 
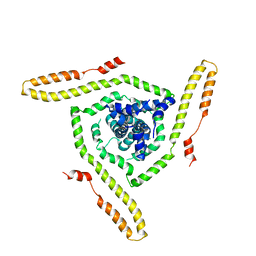 | | Model of the HIV-1 gp41 membrane-proximal external region, transmembrane domain and cytoplasmic tail | | Descriptor: | Transmembrane protein gp41 | | Authors: | Piai, A, Fu, Q, Sharp, A.K, Bighi, B, Brown, A.M, Chou, J.J. | | Deposit date: | 2021-02-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Model of the Entire Membrane-Interacting Region of the HIV-1 Fusion Protein and Its Perturbation of Membrane Morphology.
J.Am.Chem.Soc., 143, 2021
|
|
