5JPO
 
 | |
5DQS
 
 | |
5IM0
 
 | |
5Z2W
 
 | | Crystal structure of the bacterial cell division protein FtsQ and FtsB | | Descriptor: | Cell division protein FtsB, Cell division protein FtsQ, MAGNESIUM ION | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into the FtsQ/FtsB/FtsL Complex, a Key Component of the Divisome.
Sci Rep, 8, 2018
|
|
7VOV
 
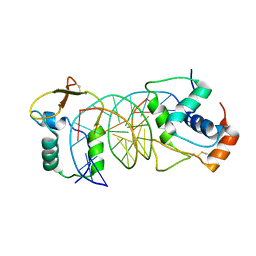 | | The crystal structure of human forkhead box protein in complex with DNA 2 | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*AP*TP*TP*TP*AP*TP*TP*AP*TP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*AP*TP*AP*AP*TP*AP*AP*AP*TP*AP*TP*T)-3'), Forkhead box protein L2 | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
7VOX
 
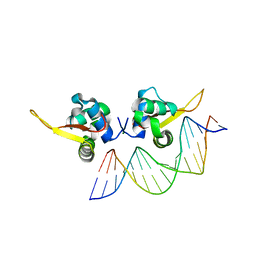 | | The crystal structure of human forkhead box protein A in complex with DNA 2 | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*AP*TP*TP*TP*AP*TP*TP*AP*TP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*AP*TP*AP*AP*TP*AP*AP*AP*TP*AP*TP*TP*T)-3'), Hepatocyte nuclear factor 3-alpha, ... | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
7VOU
 
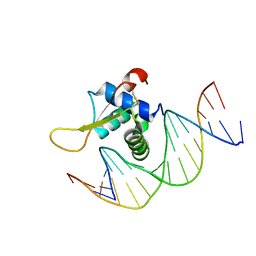 | | The crystal structure of human forkhead box protein in complex with DNA 1 | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*T)-3'), Forkhead box protein L2 | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-14 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
2KO3
 
 | | Nedd8 solution structure | | Descriptor: | NEDD8 | | Authors: | Choi, Y.S, Jeon, Y.H, Cheong, C. | | Deposit date: | 2009-09-09 | | Release date: | 2009-11-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 60th residues of ubiquitin and Nedd8 are located out of E2-binding surfaces, but are important for K48 ubiquitin-linkage.
Febs Lett., 583, 2009
|
|
7E69
 
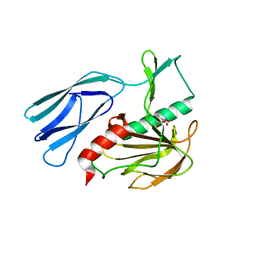 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-3 | | Descriptor: | N-oxidanyl-4-[(4-sulfamoylphenyl)methyl]benzamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E64
 
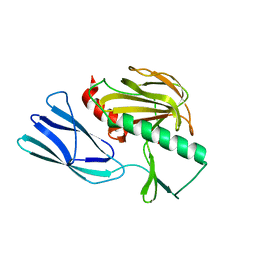 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2-2 | | Descriptor: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]amino]ethanoic acid, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E65
 
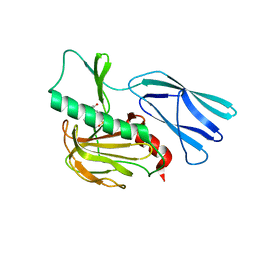 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3 | | Descriptor: | (2S)-2-acetamido-N-[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]-3-(4-sulfamoylphenyl)propanamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E67
 
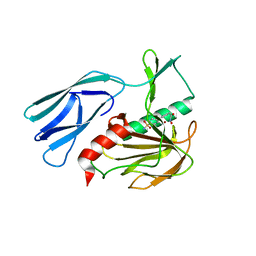 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-2 | | Descriptor: | N-oxidanyl-2-[4-(4-sulfamoylphenyl)phenyl]ethanamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E63
 
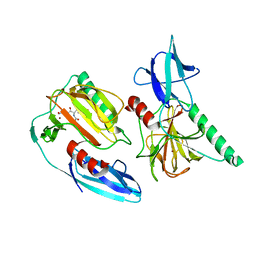 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 2-1 | | Descriptor: | 2-[[(3S)-3-acetamido-4-[[(2R)-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-4-oxidanylidene-butyl]-(cyclopentylmethyl)amino]ethanoic acid, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
7E66
 
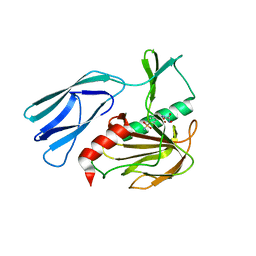 | | The crystal structure of peptidoglycan peptidase in complex with inhibitor 3-1 | | Descriptor: | N-[2-(oxidanylamino)-2-oxidanylidene-ethyl]-2-(4-sulfamoylphenyl)ethanamide, Peptidase M23, ZINC ION | | Authors: | Choi, Y, Min, K.J, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-02-21 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure-based inhibitor design for reshaping bacterial morphology
Commun Biol, 5, 2022
|
|
5H38
 
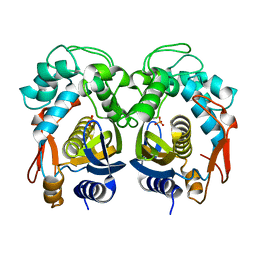 | | Structural analysis of KSHV thymidylate synthase | | Descriptor: | ORF70, PHOSPHATE ION | | Authors: | Choi, Y.M, Yeo, H.K, Park, Y.W, Lee, J.Y. | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Analysis of Thymidylate Synthase from Kaposi's Sarcoma-Associated Herpesvirus with the Anticancer Drug Raltitrexed.
PLoS ONE, 11, 2016
|
|
5H3A
 
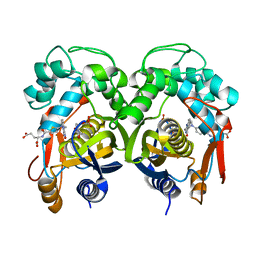 | | Structural analysis of KSHV thymidylate synthase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, ORF70, TOMUDEX | | Authors: | Choi, Y.M, Yeo, H.K, Park, Y.W, Lee, J.Y. | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Analysis of Thymidylate Synthase from Kaposi's Sarcoma-Associated Herpesvirus with the Anticancer Drug Raltitrexed.
PLoS ONE, 11, 2016
|
|
5H39
 
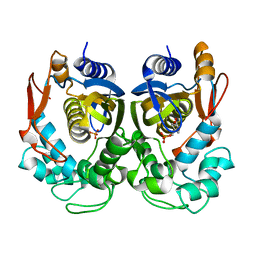 | | Structural analysis of KSHV thymidylate synthase | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, ORF70 | | Authors: | Choi, Y.M, Yeo, H.K, Park, Y.W, Lee, J.Y. | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of Thymidylate Synthase from Kaposi's Sarcoma-Associated Herpesvirus with the Anticancer Drug Raltitrexed.
PLoS ONE, 11, 2016
|
|
1QKG
 
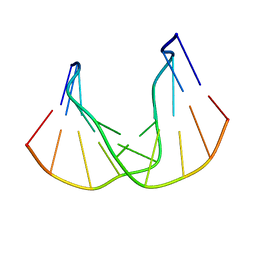 | | DNA DECAMER DUPLEX CONTAINING T-T DEWAR PHOTOPRODUCT | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*(HYD)TP*+TP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*TP*GP*CP*G)-3') | | Authors: | Lee, J.-H, Bae, S.-H, Choi, Y.-J, Choi, B.-S. | | Deposit date: | 1999-07-20 | | Release date: | 2000-05-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Dewar Photoproduct of Thymidylyl(3'-->5')-Thymidine (Dewar Product) Exhibits Mutagenic Behavior in Accordance with the "A Rule".
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2RVC
 
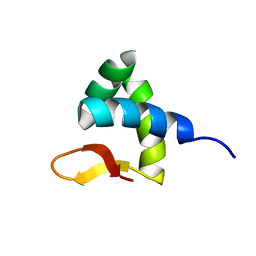 | | Solution structure of Zalpha domain of goldfish ZBP-containing protein kinase | | Descriptor: | Interferon-inducible and double-stranded-dependent eIF-2kinase | | Authors: | Lee, A, Park, C, Park, J, Kwon, M, Choi, Y, Kim, K, Choi, B, Lee, J. | | Deposit date: | 2015-07-08 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Z-DNA binding domain of PKR-like protein kinase from Carassius auratus and quantitative analyses of the intermediate complex during B-Z transition.
Nucleic Acids Res., 44, 2016
|
|
4YVM
 
 | |
5B16
 
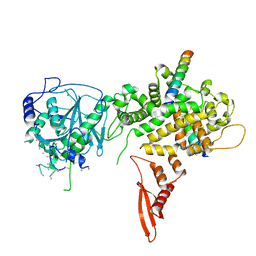 | | X-ray structure of DROSHA in complex with the C-terminal tail of DGCR8. | | Descriptor: | Microprocessor complex subunit DGCR8, Ribonuclease 3,DROSHA,Ribonuclease 3,DROSHA,Ribonuclease 3, ZINC ION | | Authors: | Kwon, S.C, Nguyen, T.A, Choi, Y.G, Jo, M.H, Hohng, S, Kim, V.N, Woo, J.S. | | Deposit date: | 2015-11-23 | | Release date: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Human DROSHA
Cell, 164, 2016
|
|
1LB4
 
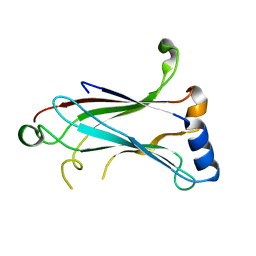 | | TRAF6 apo structure | | Descriptor: | TNF receptor-associated factor 6 | | Authors: | Ye, H, Arron, J.R, Lamothe, B, Cirilli, M, Kobayashi, T, Shevde, N.K, Segal, D, Dzivenu, O, Vologodskaia, M, Yim, M, Du, K, Singh, S, Pike, J.W, Darnay, B.G, Choi, Y, Wu, H. | | Deposit date: | 2002-04-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distinct molecular mechanism for initiating TRAF6 signalling.
Nature, 418, 2002
|
|
1LB5
 
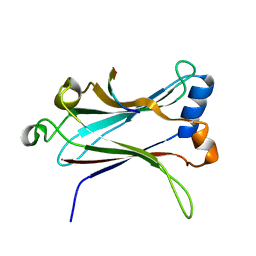 | | TRAF6-RANK Complex | | Descriptor: | TNF receptor-associated factor 6, receptor activator of nuclear factor-kappa B | | Authors: | Ye, H, Arron, J.R, Lamothe, B, Cirilli, M, Kobayashi, T, Shevde, N.K, Segal, D, Dzivenu, O, Vologodskaia, M, Yim, M, Du, K, Singh, S, Pike, J.W, Darnay, B.G, Choi, Y, Wu, H. | | Deposit date: | 2002-04-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distinct molecular mechanism for initiating TRAF6 signalling.
Nature, 418, 2002
|
|
1LB6
 
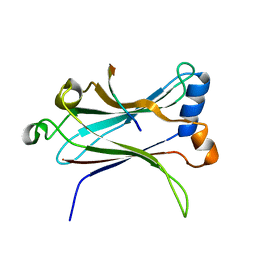 | | TRAF6-CD40 Complex | | Descriptor: | CD40 antigen, TNF receptor-associated factor 6 | | Authors: | Ye, H, Arron, J.R, Lamothe, B, Cirilli, M, Kobayashi, T, Shevde, N.K, Segal, D, Dzivenu, O, Vologodskaia, M, Yim, M, Du, K, Singh, S, Pike, J.W, Darnay, B.G, Choi, Y, Wu, H. | | Deposit date: | 2002-04-02 | | Release date: | 2002-07-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Distinct molecular mechanism for initiating TRAF6 signalling.
Nature, 418, 2002
|
|
2BD0
 
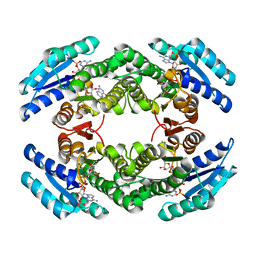 | | Chlorobium tepidum Sepiapterin Reductase complexed with NADP and Sepiapterin | | Descriptor: | BIOPTERIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, sepiapterin reductase | | Authors: | Supangat, S, Seo, K.H, Choi, Y.K, Park, Y.S, Lee, K.H. | | Deposit date: | 2005-10-19 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Chlorobium tepidum sepiapterin reductase complex reveals the novel substrate binding mode for stereospecific production of L-threo-tetrahydrobiopterin
J.Biol.Chem., 281, 2006
|
|
