3KYF
 
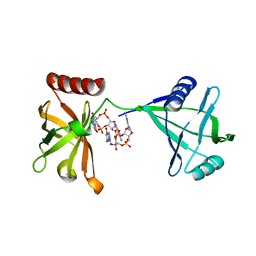 | | Crystal structure of P4397 complexed with c-di-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Putative uncharacterized protein | | Authors: | Ryu, K.S, Ko, J, Kim, H, Choi, B.S. | | Deposit date: | 2009-12-06 | | Release date: | 2010-04-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of PP4397 Reveals the Molecular Basis for Different c-di-GMP Binding Modes by Pilz Domain Proteins.
J.Mol.Biol., 398, 2010
|
|
3KYG
 
 | | Crystal structure of VCA0042 (L135R) complexed with c-di-GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Putative uncharacterized protein VCA0042 | | Authors: | Ryu, K.S, Ko, J, Kim, H, Choi, B.S. | | Deposit date: | 2009-12-06 | | Release date: | 2010-04-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of PP4397 Reveals the Molecular Basis for Different c-di-GMP Binding Modes by Pilz Domain Proteins.
J.Mol.Biol., 398, 2010
|
|
1SNH
 
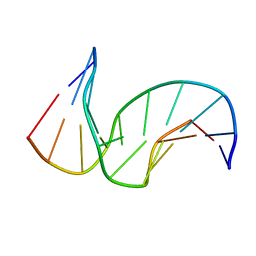 | | Solution structure of the DNA Decamer Duplex Containing Double TG Mismatches of Cis-syn Cyclobutane Pyrimidine Dimer | | Descriptor: | 5'-D(*CP*GP*CP*AP*TP*TP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*TP*GP*GP*TP*GP*CP*G)-3' | | Authors: | Lee, J.H, Park, C.J, Shin, J.S, Ikegami, T, Akutsu, H, Choi, B.S. | | Deposit date: | 2004-03-11 | | Release date: | 2004-05-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the DNA decamer duplex containing double T*G mismatches of cis-syn cyclobutane pyrimidine dimer: implications for DNA damage recognition by the XPC-hHR23B complex.
Nucleic Acids Res., 32, 2004
|
|
2JQT
 
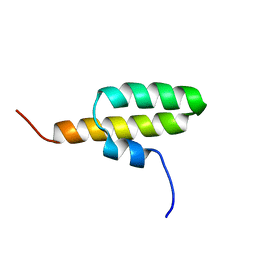 | | Structure of the bacterial replication origin-associated protein Cnu | | Descriptor: | H-NS/stpA-binding protein 2 | | Authors: | Bae, S.H, Liu, D, Lim, H.M, Lee, Y, Choi, B.S. | | Deposit date: | 2007-06-07 | | Release date: | 2008-04-22 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the nucleoid-associated protein Cnu reveals common binding sites for H-NS in Cnu and Hha.
Biochemistry, 47, 2008
|
|
4XTT
 
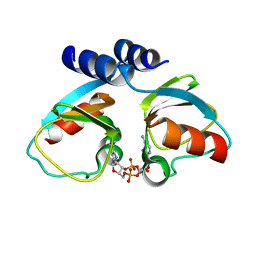 | | Structural Studies of Potassium Transport Protein KtrA Regulator of Conductance of K+ (RCK) C domain in Complex with Cyclic Diadenosine Monophosphate (c-di-AMP) | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Putative potassium transport protein | | Authors: | Kim, H, Youn, S.J, Kim, S.O, Ko, J, Lee, J.O, Choi, B.S. | | Deposit date: | 2015-01-24 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Structural Studies of Potassium Transport Protein KtrA Regulator of Conductance of K+ (RCK) C Domain in Complex with Cyclic Diadenosine Monophosphate (c-di-AMP)
J.Biol.Chem., 290, 2015
|
|
4EXT
 
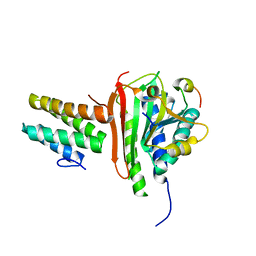 | |
1P1A
 
 | | NMR structure of ubiquitin-like domain of hHR23B | | Descriptor: | UV excision repair protein RAD23 homolog B | | Authors: | Ryu, K.S, Lee, K.J, Bae, S.H, Kim, B.K, Kim, K.A, Choi, B.S. | | Deposit date: | 2003-04-11 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a
J.Biol.Chem., 278, 2003
|
|
1PIB
 
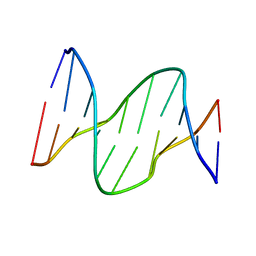 | | Solution structure of DNA containing CPD opposited by GA | | Descriptor: | 5'-D(*CP*GP*CP*AP*TP*TP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*TP*GP*AP*TP*GP*CP*G)-3' | | Authors: | Lee, J.H, Park, C.J, Shin, J.S, Choi, B.S. | | Deposit date: | 2003-05-30 | | Release date: | 2004-05-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the DNA decamer duplex containing double T*G mismatches of cis-syn cyclobutane pyrimidine dimer: implications for DNA damage recognition by the XPC-hHR23B complex.
Nucleic Acids Res., 32, 2004
|
|
1X8D
 
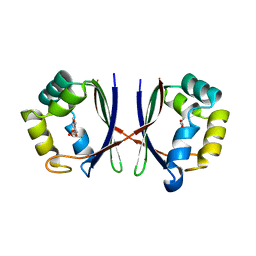 | | Crystal structure of E. coli YiiL protein containing L-rhamnose | | Descriptor: | Hypothetical protein yiiL, L-RHAMNOSE | | Authors: | Ryu, K.S, Kim, J.I, Cho, S.J, Park, D, Park, C, Lee, J.O, Choi, B.S. | | Deposit date: | 2004-08-18 | | Release date: | 2005-05-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Monosaccharide Specificity of Escherichia coli Rhamnose Mutarotase
J.Mol.Biol., 349, 2005
|
|
1YNX
 
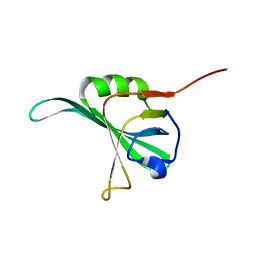 | |
1ZYH
 
 | | Structure of a Supercoiling Responsive DNA site | | Descriptor: | 5'-D(*CP*AP*AP*CP*CP*AP*GP*GP*GP*TP*TP*G)-3', 5'-D(*CP*AP*AP*CP*CP*CP*TP*GP*GP*TP*TP*G)-3' | | Authors: | Bae, S.H, Yun, S.H, Sun, D, Lim, H.M, Choi, B.S. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic basis of a supercoiling-responsive DNA element
Nucleic Acids Res., 34, 2006
|
|
1ZYF
 
 | | Structure of a Supercoiling Responsive DNA Site | | Descriptor: | 5'-D(*CP*AP*AP*CP*CP*AP*TP*GP*GP*TP*TP*G)-3' | | Authors: | Bae, S.H, Yun, S.H, Sun, D, Lim, H.M, Choi, B.S. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic basis of a supercoiling-responsive DNA element
Nucleic Acids Res., 34, 2006
|
|
1ZYG
 
 | | Structure of a Supercoiling Responsive DNA Site | | Descriptor: | 5'-D(*CP*AP*AP*CP*CP*CP*GP*GP*GP*TP*TP*G)-3' | | Authors: | Bae, S.H, Yun, S.H, Sun, D, Lim, H.M, Choi, B.S. | | Deposit date: | 2005-06-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic basis of a supercoiling-responsive DNA element
Nucleic Acids Res., 34, 2006
|
|
2MH9
 
 | |
