4R3Z
 
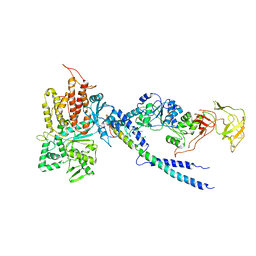 | | Crystal structure of human ArgRS-GlnRS-AIMP1 complex | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 1, Arginine--tRNA ligase, cytoplasmic, ... | | Authors: | Fu, Y, Kim, Y, Cho, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.033 Å) | | Cite: | Structure of the ArgRS-GlnRS-AIMP1 complex and its implications for mammalian translation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2ZIX
 
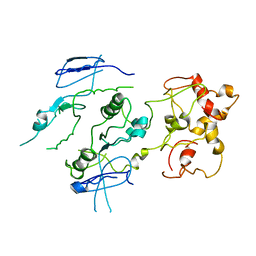 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81 | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
2ZIW
 
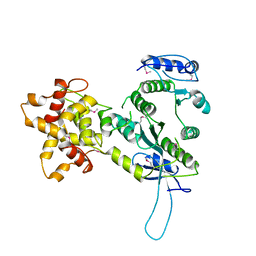 | | Crystal structure of the Mus81-Eme1 complex | | Descriptor: | Crossover junction endonuclease EME1, Mus81 protein | | Authors: | Chang, J.H, Kim, J.J, Choi, J.M, Lee, J.H, Cho, Y. | | Deposit date: | 2008-02-25 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Mus81-Eme1 complex
Genes Dev., 22, 2008
|
|
2EHO
 
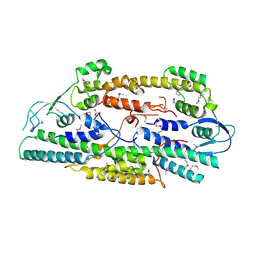 | | Crystal structure of human GINS complex | | Descriptor: | DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, GINS complex subunit 3, ... | | Authors: | Choi, J.M, Lim, H.S, Kim, J.J, Song, O.K, Cho, Y. | | Deposit date: | 2007-03-07 | | Release date: | 2007-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the human GINS complex
Genes Dev., 21, 2007
|
|
4P0P
 
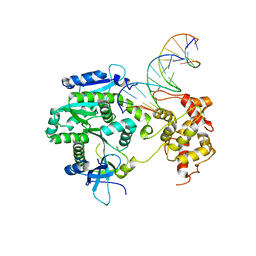 | | Crystal structure of Human Mus81-Eme1 in complex with 5'-flap DNA, and Mg2+ | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, DNA GAATGTGTGTCTCAATC, ... | | Authors: | Gwon, G.H, Baek, K, Cho, Y. | | Deposit date: | 2014-02-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates.
Embo J., 33, 2014
|
|
4P0Q
 
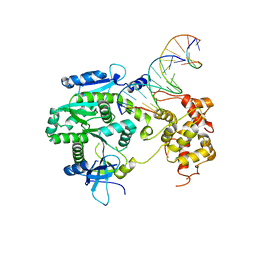 | | Crystal structure of Human Mus81-Eme1 in complex with 5'-flap DNA | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, DNA GAATGTGTGTCTCAATC, ... | | Authors: | Gwon, G.H, Baek, K, Cho, Y. | | Deposit date: | 2014-02-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates.
Embo J., 33, 2014
|
|
4P0R
 
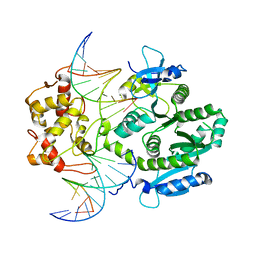 | | human Mus81-Eme1-3'flap DNA complex | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, DNA ACGTGCTTACACACAGAGGTTAGGGTGAACTT, ... | | Authors: | Gwon, G.H, Baek, K, Cho, Y. | | Deposit date: | 2014-02-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (6.501 Å) | | Cite: | Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates.
Embo J., 33, 2014
|
|
4P0S
 
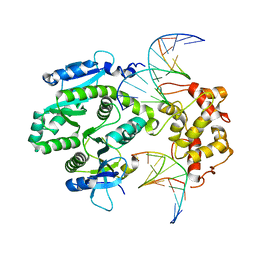 | | human Mus81-Eme1-3'flap DNA complex | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, DNA GAATGTGTGTCT, ... | | Authors: | Gwon, G.H, Baek, K, Cho, Y. | | Deposit date: | 2014-02-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates.
Embo J., 33, 2014
|
|
4BFM
 
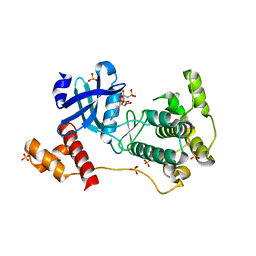 | | The crystal structure of mouse PK38 | | Descriptor: | MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION | | Authors: | Yoo, J.H, Cho, Y.S, Park, S.M, Cho, H.S. | | Deposit date: | 2013-03-21 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structures of the Kinase Domain and Uba Domain of Mpk38 Suggest the Activation Mechanism for Kinase Activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1N4M
 
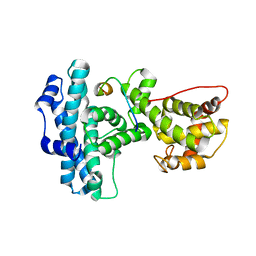 | | Structure of Rb tumor suppressor bound to the transactivation domain of E2F-2 | | Descriptor: | Retinoblastoma Pocket, Transcription factor E2F2 | | Authors: | Lee, C, Chang, J.H, Lee, H.S, Cho, Y. | | Deposit date: | 2002-10-31 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition of the E2F transactivation domain by the retinoblastoma tumor suppressor
GENES DEV., 16, 2002
|
|
1N6A
 
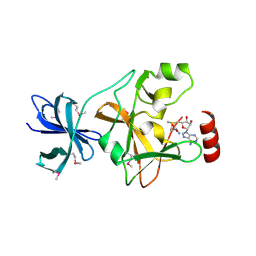 | | Structure of SET7/9 | | Descriptor: | S-ADENOSYLMETHIONINE, SET domain-containing protein 7 | | Authors: | Kwon, T.W, Chang, J.H, Kwak, E, Lee, C.W, Joachimiak, A, Kim, Y.C, Lee, J, Cho, Y. | | Deposit date: | 2002-11-09 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet
EMBO J., 22, 2003
|
|
1N6C
 
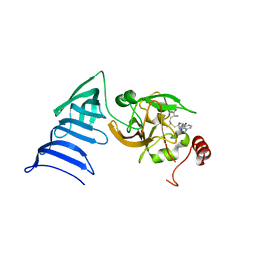 | | Structure of SET7/9 | | Descriptor: | S-ADENOSYLMETHIONINE, SET domain-containing protein 7 | | Authors: | Kwon, T.W, Chang, J.H, Cho, Y. | | Deposit date: | 2002-11-09 | | Release date: | 2003-02-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet
EMBO J., 22, 2003
|
|
3B64
 
 | |
2KLO
 
 | | Structure of the Cdt1 C-terminal domain | | Descriptor: | DNA replication factor Cdt1 | | Authors: | Khayrutdinov, B.I, Bae, W.J, Yun, Y.M, Tsuyama, T, Kim, J.J, Hwang, E, Ryu, K.-S, Cheong, H.-K, Cheong, C, Karplus, P.A, Guntert, P, Tada, S, Jeon, Y.H, Cho, Y. | | Deposit date: | 2009-07-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Cdt1 C-terminal domain: Conservation of the winged helix fold in replication licensing factors
Protein Sci., 18, 2009
|
|
2NWG
 
 | | Structure of CXCL12:heparin disaccharide complex | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Stromal cell-derived factor 1 | | Authors: | Murphy, J.W, Cho, Y, Lolis, E. | | Deposit date: | 2006-11-14 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural and Functional Basis of CXCL12 (Stromal Cell-derived Factor-1{alpha}) Binding to Heparin
J.Biol.Chem., 282, 2007
|
|
4TUG
 
 | | Crystal structure of MjMre11-DNA2 complex | | Descriptor: | DNA (5'-D(P*CP*TP*GP*TP*CP*CP*TP*AP*CP*GP*TP*GP*CP*CP*A)-3'), DNA (5'-D(P*GP*CP*AP*CP*GP*TP*AP*GP*GP*AP*CP*AP*GP*C)-3'), DNA double-strand break repair protein Mre11, ... | | Authors: | Sung, S, Cho, Y. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | DNA end recognition by the Mre11 nuclease dimer: insights into resection and repair of damaged DNA.
Embo J., 33, 2014
|
|
4TUI
 
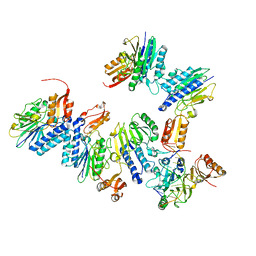 | | Crystal structure of MjMre11-DNA1 complex | | Descriptor: | DNA (5'-D(P*TP*CP*CP*TP*AP*CP*GP*TP*GP*CP*CP*AP*G)-3'), DNA (5'-D(P*TP*GP*GP*CP*AP*CP*GP*TP*AP*GP*GP*AP*C)-3'), DNA double-strand break repair protein Mre11 | | Authors: | Sung, S, Cho, Y. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | DNA end recognition by the Mre11 nuclease dimer: insights into resection and repair of damaged DNA.
Embo J., 33, 2014
|
|
5Y7Q
 
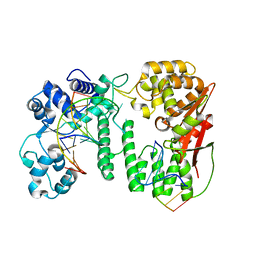 | | Crystal structure of paFAN1 bound to 2nt 5'flap DNA with gap | | Descriptor: | DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*AP*TP*TP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*CP*TP*T)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Jin, H, Cho, Y. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease.
J. Biol. Chem., 293, 2018
|
|
5Z6W
 
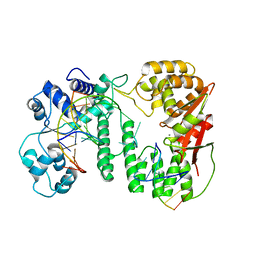 | | Crystal structure of paFAN1 bound to 2nt 5'flap DNA with gap with Manganese | | Descriptor: | DNA (5'-D(P*AP*TP*TP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*CP*TP*T)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Jin, H, Cho, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease.
J. Biol. Chem., 293, 2018
|
|
3A1J
 
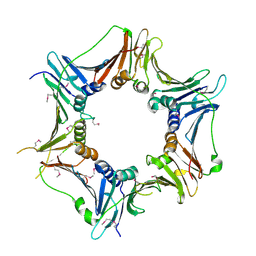 | | Crystal structure of the human Rad9-Hus1-Rad1 complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1, ... | | Authors: | Sohn, S.Y, Cho, Y. | | Deposit date: | 2009-04-08 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Human Rad9-Hus1-Rad1 Clamp
J.Mol.Biol., 390, 2009
|
|
1A77
 
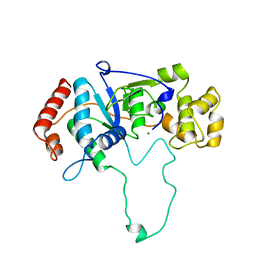 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MAGNESIUM ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
1A76
 
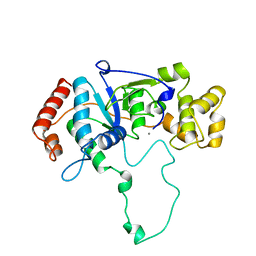 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MANGANESE (II) ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
3AUY
 
 | | Crystal structure of Rad50 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA double-strand break repair rad50 ATPase, MAGNESIUM ION | | Authors: | Lim, H.S, Cho, Y. | | Deposit date: | 2011-02-18 | | Release date: | 2011-05-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Mre11-Rad50-ATP S Complex: Understanding the Interplay between Mre11 and Rad50
To be Published
|
|
3AUZ
 
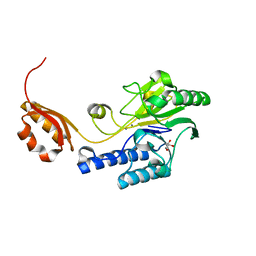 | | Crystal structure of Mre11 with manganese | | Descriptor: | DNA double-strand break repair protein mre11, GLYCEROL, MANGANESE (II) ION | | Authors: | Park, Y.B, Cho, Y. | | Deposit date: | 2011-02-18 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Crystal Structure of the Mre11-Rad50-ATP S Complex: Understanding the Interplay between Mre11 and Rad50
To be Published
|
|
3AUX
 
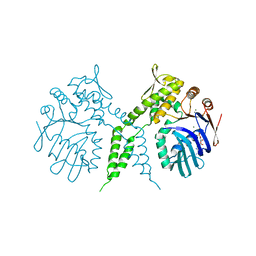 | | Crystal structure of Rad50 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA double-strand break repair rad50 ATPase, MAGNESIUM ION | | Authors: | Lim, H.S, Cho, Y. | | Deposit date: | 2011-02-17 | | Release date: | 2011-05-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Mre11-Rad50-ATP S Complex:Understanding the Interplay between Mre11 and Rad50
To be Published
|
|
