7LUM
 
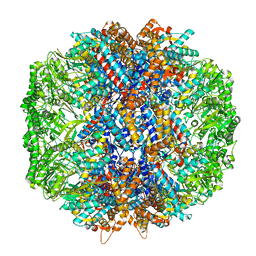 | | Human TRiC in ATP/AlFx closed state | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Knowlton, J.J, Gestaut, D, Ma, B, Taylor, G, Seven, A.B, Leitner, A, Wilson, G.J, Shanker, S, Yates, N.A, Prasad, B.V.V, Aebersold, R, Chiu, W, Frydman, J, Dermody, T.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural and functional dissection of reovirus capsid folding and assembly by the prefoldin-TRiC/CCT chaperone network.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LJY
 
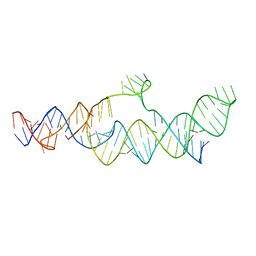 | | Cryo-EM structure of the B dENE construct complexed with a 28-mer poly(A) | | Descriptor: | B dENE construct, poly(A) | | Authors: | Torabi, S, Chen, Y, Zhang, K, Wang, J, DeGregorio, S, Vaidya, A, Su, Z, Pabit, S, Chiu, W, Pollack, L, Steitz, J. | | Deposit date: | 2021-02-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural analyses of an RNA stability element interacting with poly(A).
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WLN
 
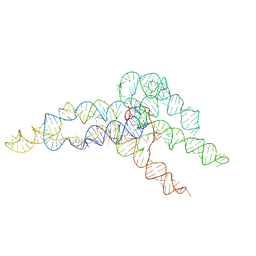 | | hc16 ligase product models, 10.0 Angstrom resolution | | Descriptor: | RNA (349-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLL
 
 | | Apo F. nucleatum glycine riboswitch models, 10.0 Angstrom resolution | | Descriptor: | RNA (171-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLJ
 
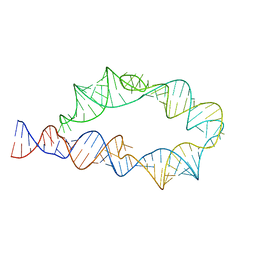 | | ATP-TTR-3 with AMP models, 9.6 Angstrom resolution | | Descriptor: | RNA (130-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLK
 
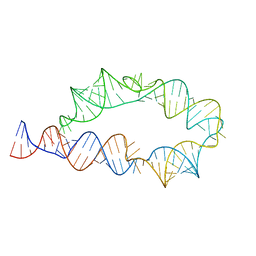 | | Apo ATP-TTR-3 models, 10.0 Angstrom resolution | | Descriptor: | RNA (130-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLT
 
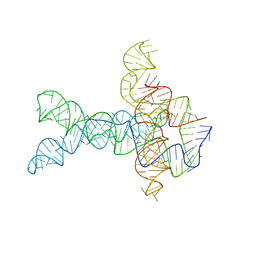 | | Apo V. cholerae glycine riboswitch models, 4.8 Angstrom resolution | | Descriptor: | RNA (231-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
3J03
 
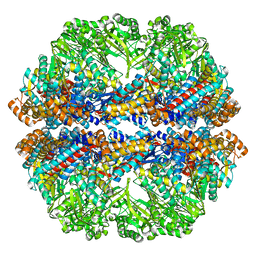 | | Lidless Mm-cpn in the closed state with ATP/AlFx | | Descriptor: | Lidless Mm-cpn | | Authors: | Zhang, J, Ma, B, DiMaio, F, Douglas, N.R, Joachimiak, L, Baker, D, Frydman, J, Levitt, M, Chiu, W. | | Deposit date: | 2011-02-10 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure.
Structure, 19, 2011
|
|
3J40
 
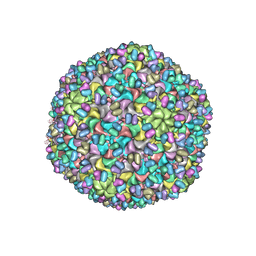 | | Validated Near-Atomic Resolution Structure of Bacteriophage Epsilon15 Derived from Cryo-EM and Modeling | | Descriptor: | gp10, gp7 | | Authors: | Baker, M.L, Hryc, C.F, Zhang, Q, Wu, W, Jakana, J, Haase-Pettingell, C, Afonine, P.V, Adams, P.D, King, J.A, Jiang, W, Chiu, W. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6WLU
 
 | | V. cholerae glycine riboswitch with glycine models, 5.7 Angstrom resolution | | Descriptor: | RNA (231-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLQ
 
 | | Apo SAM-IV riboswitch models, 4.7 Angstrom resolution | | Descriptor: | RNA (119-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLO
 
 | | hc16 ligase models, 11.0 Angstrom resolution | | Descriptor: | RNA (338-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
3J7W
 
 | | Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions | | Descriptor: | Major capsid protein 10A | | Authors: | Guo, F, Liu, Z, Fang, P.A, Zhang, Q, Wright, E.T, Wu, W, Zhang, C, Vago, F, Ren, Y, Jakata, J, Chiu, W, Serwer, P, Jiang, W. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3J7X
 
 | | Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions | | Descriptor: | Major capsid protein 10A | | Authors: | Guo, F, Liu, Z, Fang, P.A, Zhang, Q, Wright, E.T, Wu, W, Zhang, C, Vago, F, Ren, Y, Jakata, J, Chiu, W, Serwer, P, Jiang, W. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3J7V
 
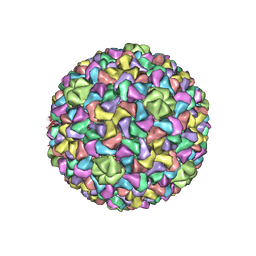 | | Capsid Expansion Mechanism Of Bacteriophage T7 Revealed By Multi-State Atomic Models Derived From Cryo-EM Reconstructions | | Descriptor: | Major capsid protein 10A | | Authors: | Guo, F, Liu, Z, Fang, P.A, Zhang, Q, Wright, E.T, Wu, W, Zhang, C, Vago, F, Ren, Y, Jakata, J, Chiu, W, Serwer, P, Jiang, W. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3J0G
 
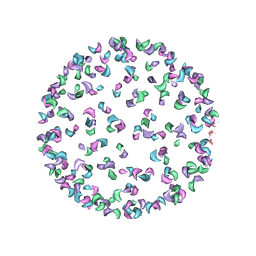 | |
3J0C
 
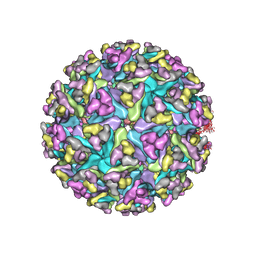 | | Models of E1, E2 and CP of Venezuelan Equine Encephalitis Virus TC-83 strain restrained by a near atomic resolution cryo-EM map | | Descriptor: | Capsid protein, E1 envelope glycoprotein, E2 envelope glycoprotein | | Authors: | Zhang, R, Hryc, C.F, Cong, Y, Liu, X, Jakana, J, Gorchakov, R, Baker, M.L, Weaver, S.C, Chiu, W. | | Deposit date: | 2011-06-22 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | 4.4 A cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus.
Embo J., 30, 2011
|
|
6UET
 
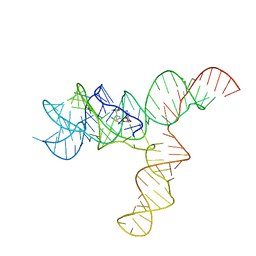 | | SAM-bound SAM-IV riboswitch | | Descriptor: | RNA (119-MER), S-ADENOSYLMETHIONINE | | Authors: | Zhang, K, Li, S, Kappel, K, Pintilie, G, Su, Z, Mou, T, Schmid, M, Das, R, Chiu, W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 angstrom resolution.
Nat Commun, 10, 2019
|
|
6UES
 
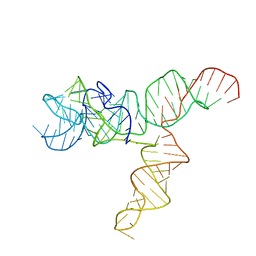 | | Apo SAM-IV Riboswitch | | Descriptor: | RNA (119-MER) | | Authors: | Zhang, K, Li, S, Kappel, K, Pintilie, G, Su, Z, Mou, T, Schmid, M, Das, R, Chiu, W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 angstrom resolution.
Nat Commun, 10, 2019
|
|
6V5C
 
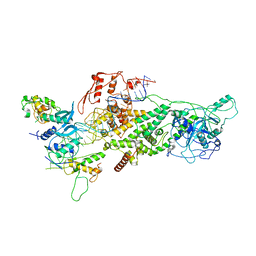 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - partially docked state | | Descriptor: | Microprocessor complex subunit DGCR8, Pri-miR-16-2 (66-MER), Ribonuclease 3 | | Authors: | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
6V5B
 
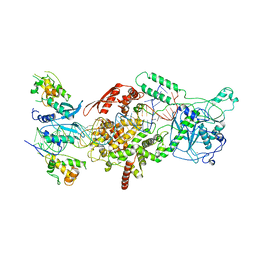 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - Active state | | Descriptor: | CALCIUM ION, Microprocessor complex subunit DGCR8, Pri-miR-16-2 (78-MER), ... | | Authors: | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
6VQV
 
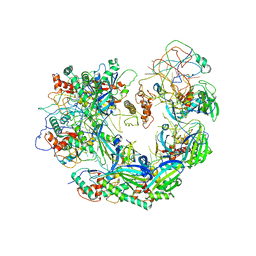 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF9 | | Descriptor: | AcrF9, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VQW
 
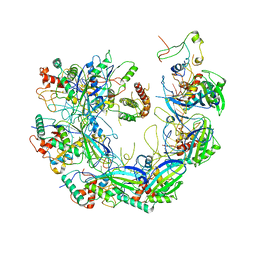 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF8 | | Descriptor: | AcrF8, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VQX
 
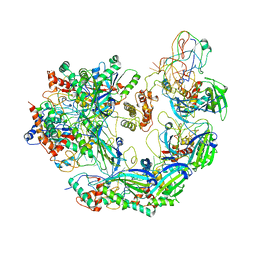 | | Type I-F CRISPR-Csy complex with its inhibitor AcrF6 | | Descriptor: | AcrF6, CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Zhu, Y, Huang, Z, Chiu, W. | | Deposit date: | 2020-02-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Inhibition mechanisms of AcrF9, AcrF8, and AcrF6 against type I-F CRISPR-Cas complex revealed by cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3IZJ
 
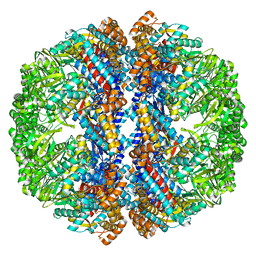 | | Mm-cpn rls with ATP and AlFx | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
